Incision enzyme mediated real-time multiple cross nucleic acid displacement amplification technology and application
A technology of restriction endonucleases and amplification primers, which can be used in the determination/testing of microorganisms, biochemical equipment and methods, etc., and can solve problems such as expensive, time-consuming and labor-consuming, and poor diagnostic sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
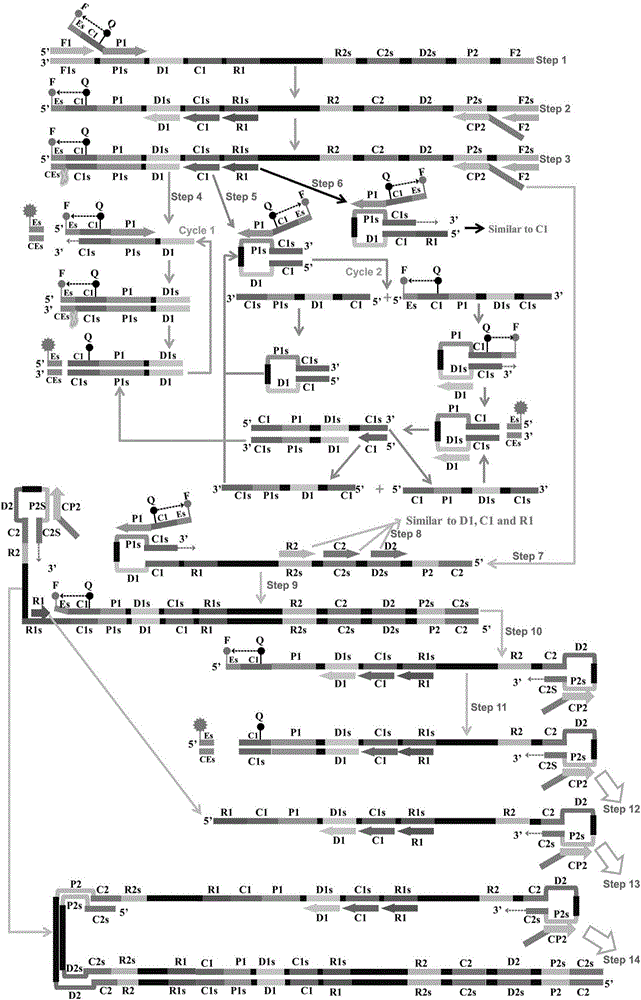
[0065] Embodiment 1. Standard ET-MCDA detects
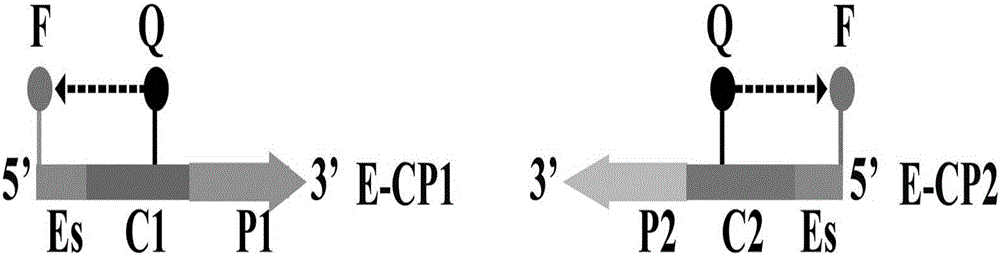
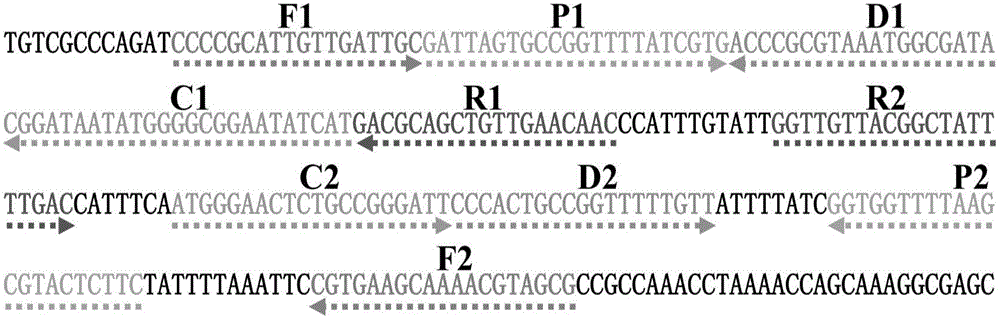
[0066] 1.1 Establish a standard ET-MCDA reaction system: the concentration of cross primers E-CP1 and CP1 is 30pmol, the concentration of cross primer CP2 is 60pmol, the concentration of replacement primers F1 and F2 is 10pmol, the concentration of amplification primers R1, R2, D1 and D2 The concentration is 30pmol, the concentration of amplification primers C1 and C2 is 20pmol, 12.5μL 2×Mix buffer reaction solution, 10U strand displacement DNA polymerase (Bst), 15U restriction endonuclease (Nb.BsrDI), 1μL template , add deionized water to 25 μl. The entire reaction was kept at 63°C for 1 hour, and the reaction was terminated at 80°C for 5 minutes.
[0067] 1.2 Prove the feasibility of the designed MCDA amplification reaction
[0068] Visible color change method: Under standard ET-MCDA reaction conditions, ET-MCDA generates a large amount of pyrophosphate ions when amplifying DNA, which can capture manganese ions bound to calce...
Embodiment 2
[0075] Embodiment 2. Multiple ET-MCDA detection
[0076] In order to obtain stable multiple fluorescence detection patterns, the standard ET-MCDA system was optimized to adapt to multiple detection, and a multiple detection system was established. Under the multiplex detection system, we added two sets of primers and corresponding templates to the multiplex reaction mixture at the same time, detected by a fluorescence detector, and obtained a stable multiplex fluorescence detection map, as shown in Figure 7 (signals of group A and group B are generated simultaneously, From the same reaction tube. Group A signals come from the Cy5 signal channel, representing the detection of Salmonella, signals 1 to 12 represent the amount of detection samples are 25ng, 2.5ng, 250pg, 25pg, 2.5pg, 250fg, 25fg, 12.5fg, 6.25 fg, 3.125fg, 1.56fg to 0.78fg / reaction tube, signal 13 represents the negative control; group B signals come from the HEX channel, representing the detection of Shigella, sig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com