Processes to prepare elongated 2-ketoacids and c6-c10 compounds therefrom via genetic modifications to microbial metabolic pathways
A C6-C10, genetic modification technology, applied in the field of engineering enzymes, can solve problems such as non-octanol and small n-octanol
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Modified LeuA (ie, "LeuA'") enzymes are prepared.
[0036] This example represents one embodiment for making an engineered LeuA' enzyme. This is accomplished by starting with an E. coli organism transformed with a plasmid containing a modified LeuA gene to generate a 2-isopropylmalate synthase variant (LeuA') for capturing the 2-ketoacid of interest for catalysis, the 2-isopropylmalate synthase variant has a higher than average catalytic efficiency (K cat / K M ). For this purpose, the specific gene identified as suitable is LeuA; GenBank Accession No. NC_000913.3 Gene ID: 947465.
[0037] In formula K cat / K M Medium, K cat It is "conversion number", unit = sec -1 ;K M is the Michaelis-Menten constant; the conversion number is equal to VMax / [E], where V Max is the maximum speed, [E] is the enzyme concentration. This equation, applied to reactions obeying Michaelis-Menten kinetics, generally provides the amount (in moles) of substrate converted to product in 1 ...
Embodiment 2
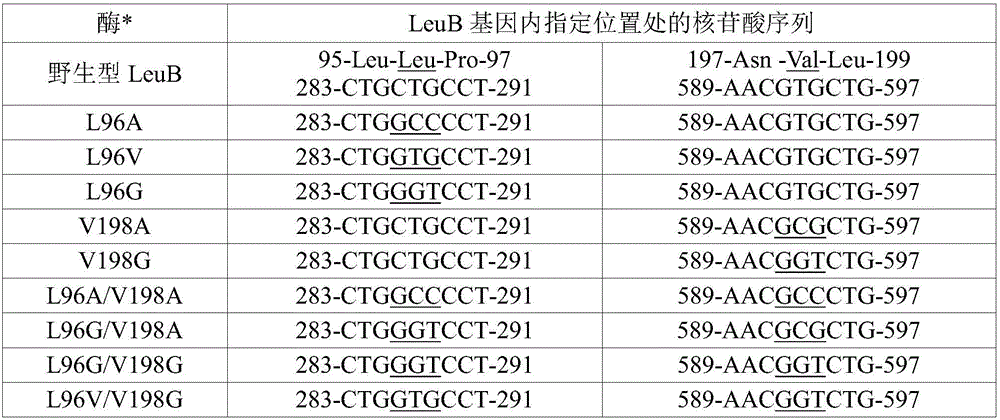
[0041] I. Preparation of E. coli LeuB' (isopropylmalate dehydrogenase) variants with increased activity towards 3-hexylmalate (3-HM).
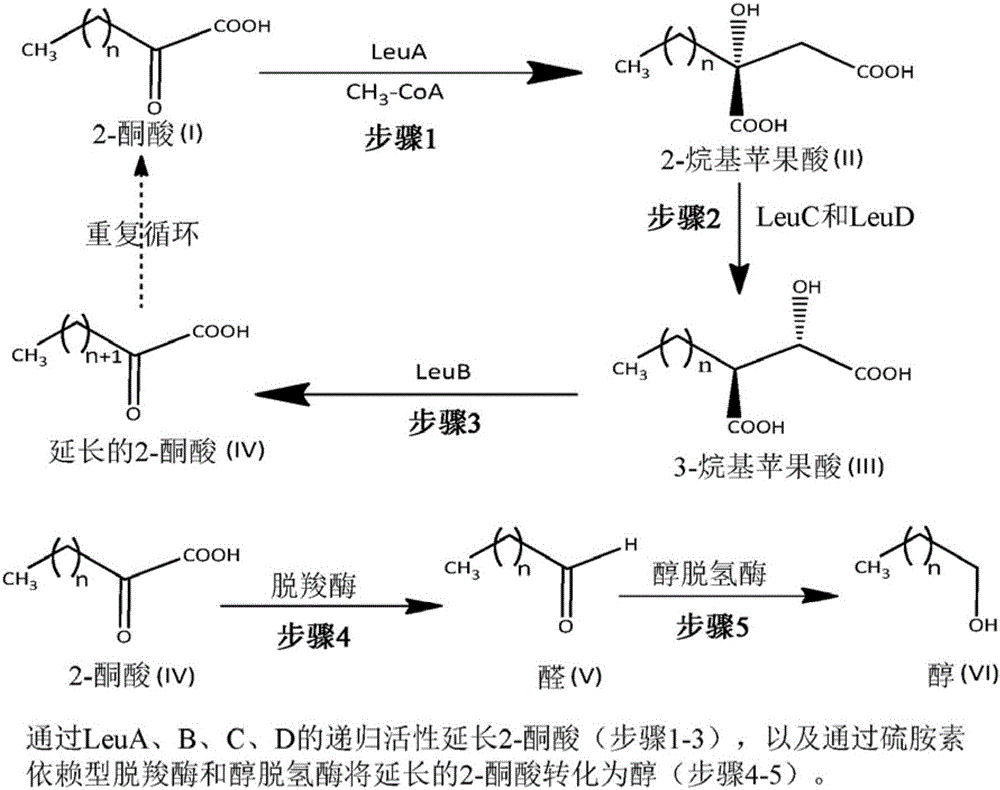
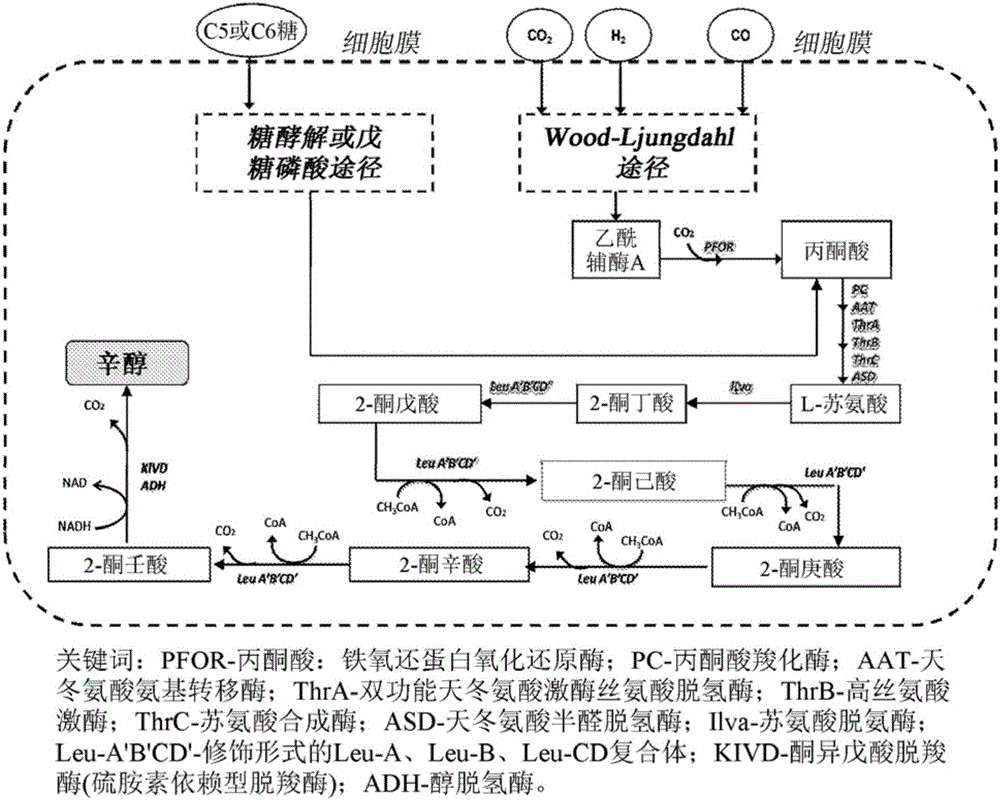
[0042] During the biosynthesis of 2-ketononanoic acid via the recursive activity of the LeuABCD pathway, 3-alkylmalates of various lengths are formed as substrates for LeuB. For efficient biosynthesis of 2-ketononanoic acid, LeuB is expected to efficiently capture 3-ethylmalate (intermediate III, n=2; figure 1 ), 3-propyl malic acid (intermediate III, n=3; figure 1 ), 3-butylmalic acid (3-BM; Intermediate III, n=4; figure 1 ), 3-amylmalic acid (intermediate III, n=5; figure 1 ) and 3-hexylmalic acid (3-HM; Intermediate III, n=6; figure 1 ) for catalysis. Native LeuB is relatively inefficient at capturing longer unnatural 3-alkylmalate substrates. In order to improve the efficiency of native LeuB in capturing 3-hexylmalate for catalysis, the active site of native LeuB was modified using protein engineering techniques described below. ...
Embodiment 3
[0084] Modified LeuCD (ie, "LeuCD'") enzymes are prepared.
[0085] In this embodiment of Example 3, a method similar to that used in Example 1 for the LeuA' enzyme is described. The method begins with an E. coli organism transformed with a plasmid containing the LeuC and LeuD genes, resulting in the production of a modified 2-isopropylmalate isomerase complex that complexes body in the 2-alkylmalic acid ( figure 1 Among them, n=1-5 in the intermediate II) isomerized to their corresponding 3-alkyl malic acid ( figure 1 In intermediate III, n=1-5) has higher catalytic efficiency (k cat / K M ). By changing some genetic codes of the LeuC and LeuD genes (LeuC: GenBank Accession No. NC000913.3 Gene ID: 945076; and LeuD: GenBank Accession No. NC000913.3, Gene ID: 945642) obtained from GenBank, that is, substituting the amino acid sequence One or more amino acids, synthesize the modified LeuC and LeuD genes. Each engineered variant was prepared by substituting one or more res...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com