Oligonucleotide probe
An oligonucleotide probe and probe technology, applied in the field of oligonucleotide probes, can solve problems such as high application threshold, high cost, and difficulty in wide-scale promotion, and achieve stable probes, simple preparation, and wide application range wide effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
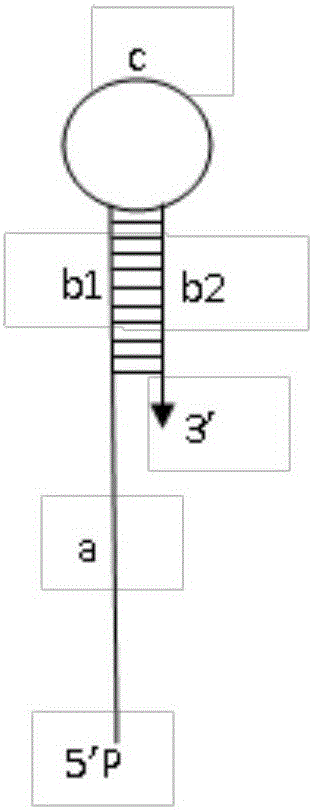
[0044] Embodiment 1, with reference to attached figure 1 , design PCR primers and probes for a synthetic target nucleic acid molecule.
[0045] Synthetic target sequence T1:
[0046] 5'- TTAGTTAAT CGGCGGGTTTTCGACGGGAAT -3'
[0047] Front primer (T1-F):
[0048] 5'-P-AAATACGAACCAAAACGCTCCCC-3'
[0049] Rear primer (T1-R):
[0050] 5'-TTATATGTCGGTTACGTGCGTTTATAT-3'
[0051] Probe (P1):
[0052] 5'-P- ATTCCCGTCGAAAACCCGCCG AAA AAA-3'
[0053] The front primer (T1-F) corresponds to the part in bold in the template (T1), the back primer (T1-R) corresponds to the italic part of T1, and the structure of the probe (P1) consists of a sequence complementary to the template sequence (underlined) , with figure 1 -a), stem sequence (marked in black box, appended figure 1 -b1, b2), ring sequence (the part between the two black boxes, attached figure 1 -c) consists of three parts, wherein the italic bold part is the G-quadruplex sequence introduced by the probe, which...
Embodiment 2
[0054] Embodiment 2, single-stranded target nucleic acid detection example
[0055] The probe designed in Example 1 was used to detect the single-stranded target nucleic acid T1.
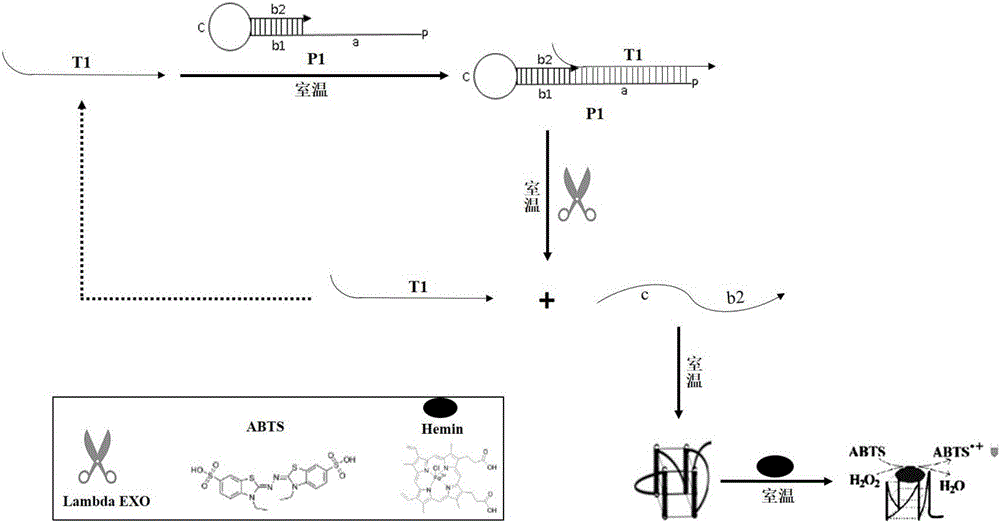
[0056] Refer to attached figure 2 The principle of single-stranded DNA detection shown is to add 2.5ul 10×Taqbuffer, 0.3uMT1, 1μMP1, 108mM NaCl solution to the 25ul color reaction system in sequence, then add 1ul Lambda enzyme, digest at 37°C for 30min, and finally add 1uM Hemin, 2mMABTS, 1mM H 2 o 2 . A negative control is: system without T1. Observe the color change with the naked eye or measure the absorbance at a wavelength of 414nm with a microplate reader. The color results are attached Figure 4 As shown, when T1 is not included in the system, no color signal visible to the naked eye is produced, and when T1 exists in the system, it can be observed that the reaction solution is obviously green. attached Figure 5 For the measurement result of chromogenic absorption value, when the con...
Embodiment 3
[0057] Embodiment 3, double-stranded target nucleic acid detection example
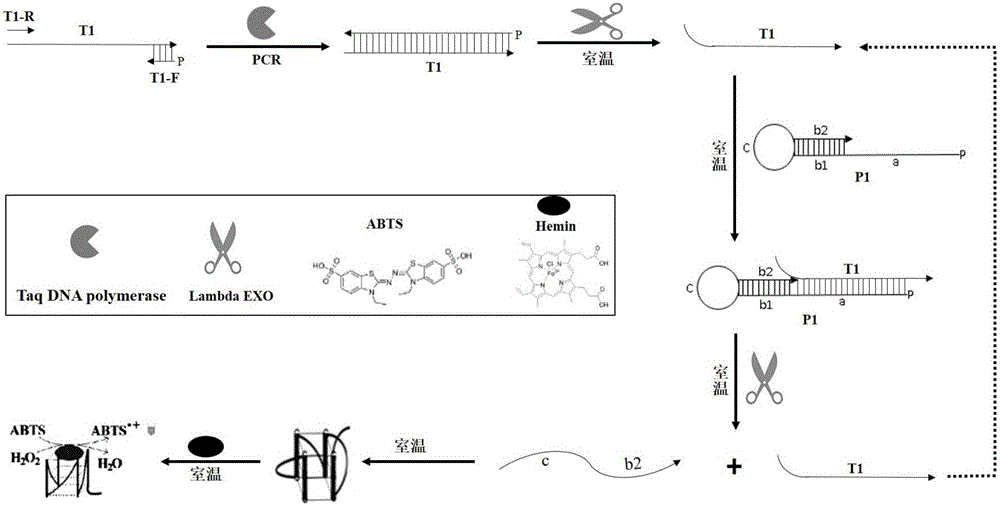
[0058] Use T1 as a template to perform PCR to form a double-stranded nucleic acid, and use the primers and probes designed in Example 1 to detect the PCR double-stranded nucleic acid product.
[0059] Refer to attached image 3 The principle of double-stranded DNA detection shown above is to firstly take a small amount of T1 for ordinary PCR to obtain a large amount of double-stranded products containing 5'-phosphorylated T1 complementary strands, and then add probes and Lambda enzymes to the PCR system to digest and release G- Quadruplex reporter molecules, and finally display the results by color reaction.
[0060] (1) PCR reaction system and reaction conditions
[0061]
[0062] The PCR conditions were: 94°C pre-denaturation for 2 minutes; 94°C denaturation for 30s, 60°C renaturation for 45s, 72°C extension for 45s, 12 cycles.
[0063] A negative control is: system without T1.
[0064] (2) A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com