Multiple improved orange/red fluorescence protein
A red fluorescent protein and protein technology, which is applied in the field of improved orange/red fluorescent protein, can solve the problems of not being able to meet the requirements of confocal imaging, shortening of photostability, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] The construction of embodiment 1.DsRed mutant library:
[0029] Through the analysis of the protein structure of DsRed (see SEQ.ID.NO: 1 for the amino acid sequence) by Discoverystudio4.0 software, find out 48 sites that may affect the structure of DsRed, these 48 sites are: R2, S4, K5, E10 , V16, R17, T21, E32, R36, H41, N42, V44, K47, Q64, F65, Q66, V71, V73, K83, L85, F91, E99, C117, F118, F124, I125, V127, T147, L150 , R153, V156, E160, I161, H162, K163, A164, L174, V175, F177, S179, I180, Y192, Y194, S197, I210, T217, G219, L225, stagger these 48 points into 12 groups (Table 2), performing saturation mutations on the amino acids at each site in each group, so that 12 groups of saturation mutation gene pools were obtained; in addition, the DsRed gene was randomly mutated by the error-prone PCR method to obtain a group of mutation gene pools; 13 groups of mutated genes were randomly recombined by DNAShuffling method, inserted into pQE30 vector, and pQE30-DsRed-DS li...
Embodiment 2
[0035] Screening of embodiment 2.DsRed mutant clones:
[0036] Spread the DsRed-DS library evenly on the ZYM-AT self-inducing medium plate (1% tryptone, 0.5% yeast extract, 25mM Na 2 HPO 4 , 25mMKH 2 PO 4 , 50mM NH 4 Cl, 5mM Na 2 SO 4 , 0.5% glycerol, 0.05% glucose, 0.2% a-lactose, 2mM MgSO 4 , 0.2x trace elements, 1% agar and 50 μg / mL Amp), cultured overnight at 30°C, the single clones in the DsRed-DS library expressed mutant fluorescent proteins, showing different brightness and colors. Observe the plate and circle the brighter red colonies with a marker. The next step is to prepare ZYM-AT self-induction liquid medium, and add 200 μl ZYM-AT liquid medium to each well of a 96-well deep-well plate. Then these bright red colonies were picked out and inoculated into 96-well plates, a total of 1820 clones were picked, and all deep-well plates were placed in a shaker at 30°C and 200rpm for 16 hours for fluorescent protein expression. The overnight expression bacterial sol...
Embodiment 3
[0040] Construction and expression of embodiment 3.sRFP mutant fluorescent protein and mOrange2 fluorescent protein eukaryotic expression vector:
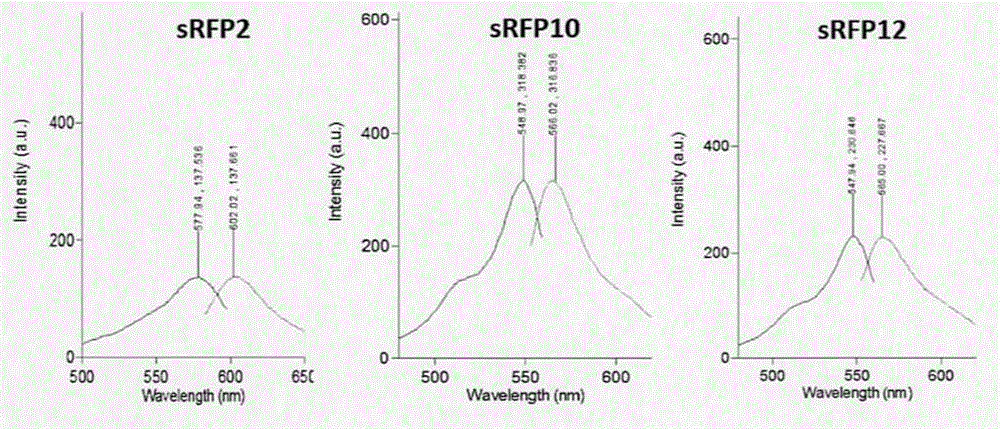
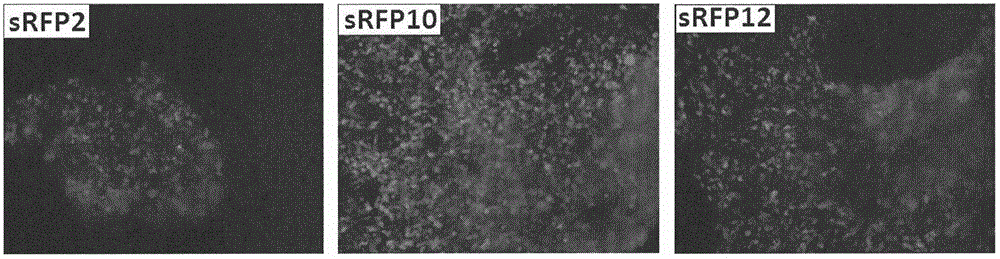
[0041]Use the primers sRFP-inf-F: 5'GGTACCGCTAGCGGATCCATGGATAGCACTGAGAACGTCA3' (SEQ.ID.NO: 10) and sRFP-inf-R: 5'GGCCGCCTCTAGACTCGAGCTACTGGAACAGGTGGTGGC3' (SEQ.ID.NO: 11) to use these three mutants as templates respectively The sRFP2, sRFP10 and sRFP12 genes were amplified and inserted into the eukaryotic expression vector pCMV3 by infusion enzymes, and the correct plasmids pCMV3-sRFP2, pCMV3-sRFP10 and pCMV3-sRFP12 were identified. Purify the pCMV3-sRFP2, pCMV3-sRFP10 and pCMV3-sRFP12 plasmids with transfection grade purity to transfect 293H cells. The specific steps are: 293H cells were cultured in DMEM medium containing 10% fetal bovine serum at 37°C and 5% carbon dioxide. Before transfection, cells were cultured at 1×10 5 The density of cells / well was seeded in a 24-well plate, and used for transfection after 24 hours of cultu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com