Method for constructing double-stem-loop structure DNA template to detect nucleic acid based on ligation reaction
A ligation reaction and template detection technology, applied in the field of biological analysis, can solve problems such as difficult to distinguish single base differences, false positive amplification signals of primer dimers, specificity difficult to meet mutations, etc., and achieve the goal of reducing cross-contamination of reagents Risk, Avoidance of Radiation Hazards, Effect of Improving Sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
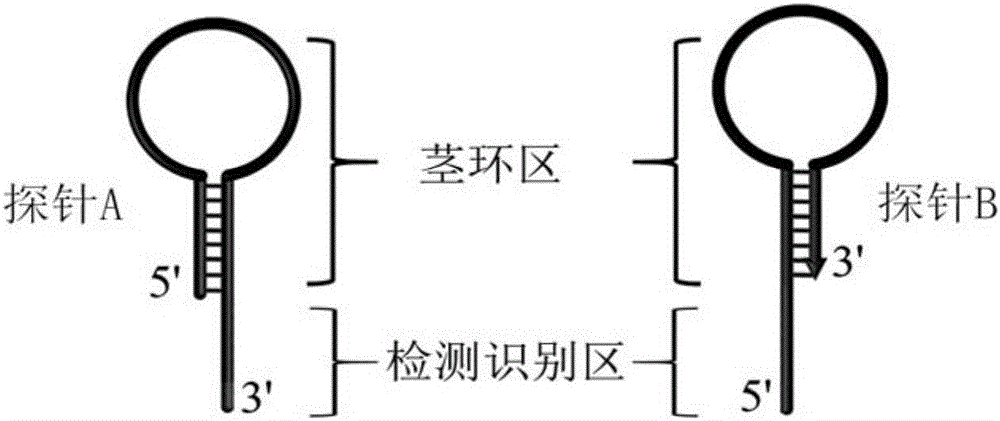
[0027] Taking let-7a as the microRNA analysis model, according to the let-7a sequence 5′-UGAGGUAGUAGGUUGUAUAGUU-3′, probe A and probe B with stem-loop structure were designed and synthesized. The base sequence of probe A is 5′-CGACAGCAGAGGATTTGTTGTGTGGAAGTGTGAGCGGATTTTCCTCTGCTGTCGTTTTAACTATACAAC- 3' (provided by Treasure Bioengineering (Dalian) Co., Ltd.); the base sequence of probe B is 5'-CTACTACCTCATTTTATCGTCGTGACTGTTTGTAATAGGACAGAGCCCCGCACTTTTCAGTCACGACGAT-3' (provided by Treasure Bioengineering (Dalian) Co., Ltd.), wherein the 5' of probe B Modified phosphate groups. Schematic diagram of the structure of probe A and probe B as figure 1 shown.
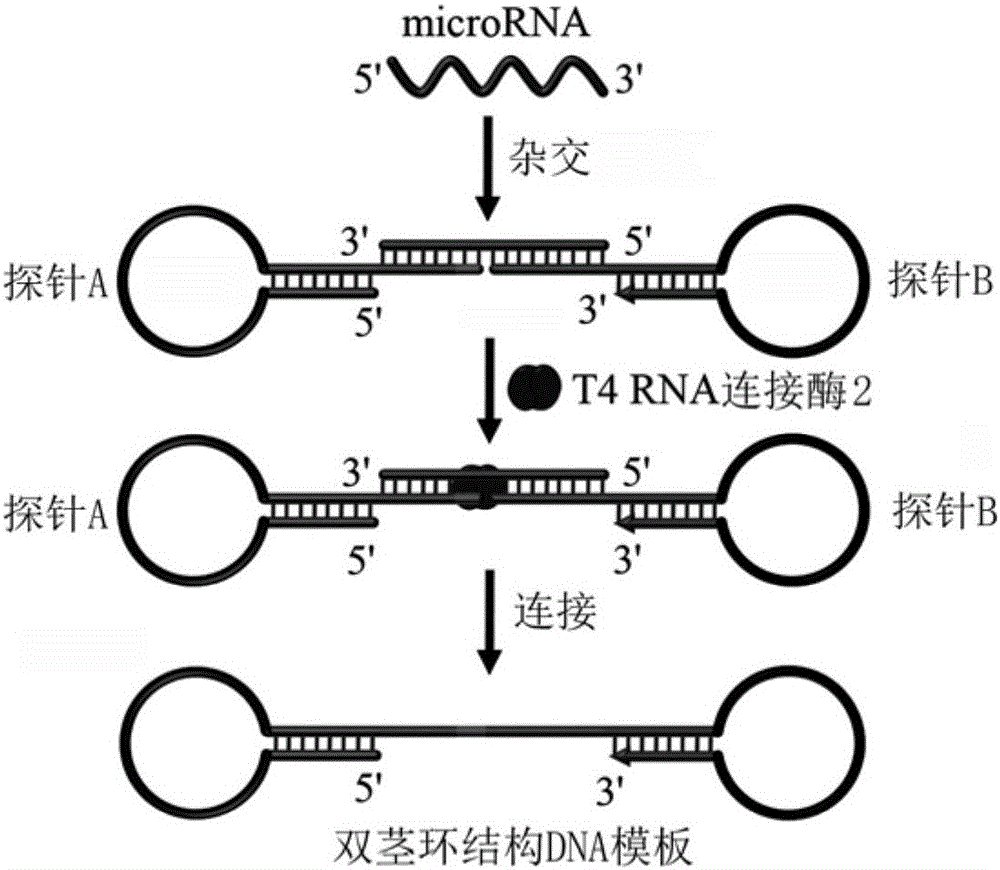
[0028] Such as figure 2 As shown, when there is let-7a, the detection and recognition regions of probe A and probe B are specifically hybridized with let-7a respectively, using let-7a as a template, under the catalysis of T4RNA ligase 2, probe A and probe B Probe B is connected to form a double-stem-loop structure DNA, which ca...
Embodiment 2
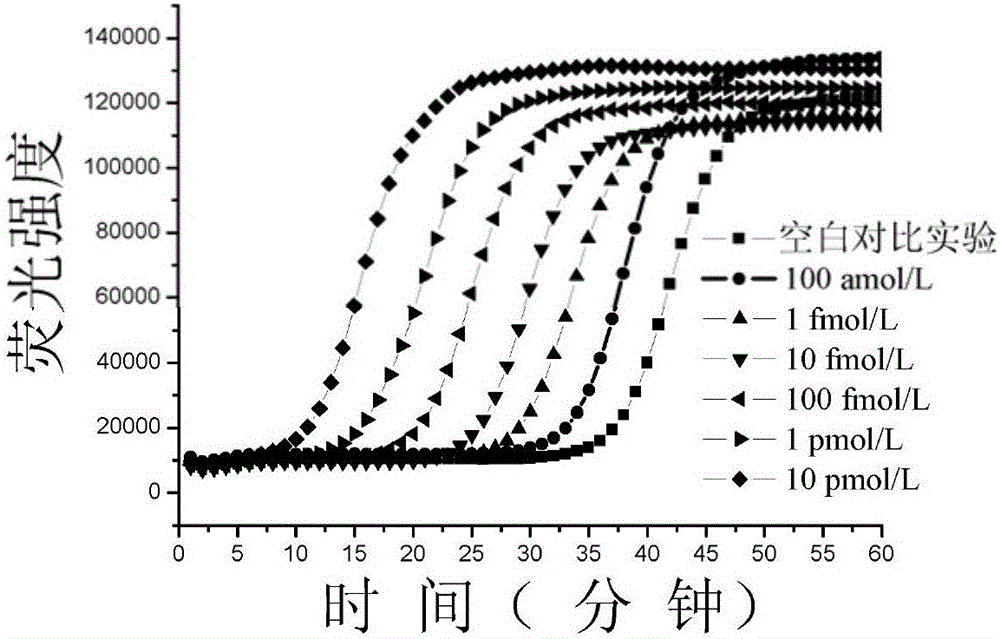
[0035] The human let-7miRNA family is a highly homologous similar sequence with a difference of only one or several bases. Use probe A and probe B designed for let-7a in Example 1 to detect let-7b (5'-UGAGGUAGUAGGUUGUGUGGUU-3') and let-7c (5'-UGAGGUAGUAGGUUGUAUGGUU-3') in the let-7 family , let-7d (5′-AGAGGUAGUAGGUUGCAUAGUU-3′), let-7e (5′-UGAGGUAGGAGGUUGUAUAGUU-3′), the detection method is to use let-7b, let-7c, let-7d, let- 7e to replace let-7a, other experimental steps were the same as in Example 1, and the real-time fluorescence curves were measured respectively. Let-7b and let-7d differ from the let-7a sequence by two bases, and let-7c and let-7e differ from the let-7a sequence by one base. Test results such as Figure 5 shown. Depend on Figure 5 It can be seen that let-7a can be clearly distinguished from other miRNAs in the let-7 family, the non-specific interference of let-7b is 4.6%, the non-specific interference of let-7c is 12.9%, the non-specific interference ...
Embodiment 3
[0037] Taking the mutant DNA fragment of exon 8 of the human P53 gene as an example, the mutant DNA (5′-TGTTTGTGCCTGTCCTGGGAGAGACTGGCGCACAGAGGAAGAGAATCTC-3′) was used as the detection target sequence, and probe A and probe B with a stem-loop structure were designed and synthesized. The base sequence of needle A is 5′-CGACAGCAGAGGATTTGTTGTGTGGAAGTGTGAGCGGATTTTCCTCTGCTGTCGCTCTTCCTCTGTGCGCCA-3′ (provided by Bao Bioengineering (Dalian) Co., Ltd.); the base sequence of probe B is 5′-GTCTCTCCCAGGACAGGCATCGTCGTGACTGTTTGTAATAGGACAGAGCCCCGCACTTTCAGTCACGACGAT-3′ (provided by Bao Bioengineering (Dalian) Co., Ltd.), wherein the 5' end of probe B is modified with a phosphate group.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com