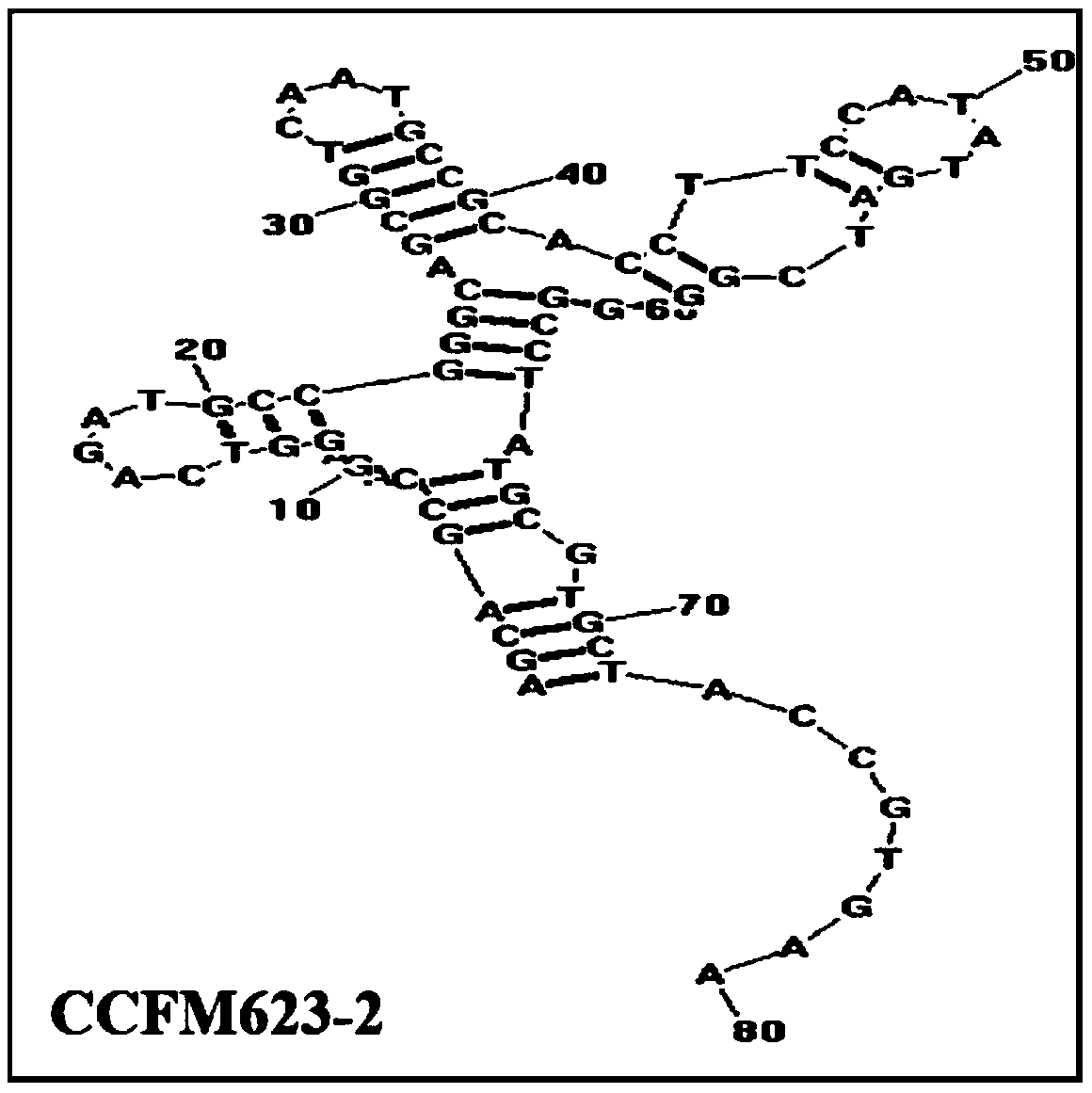
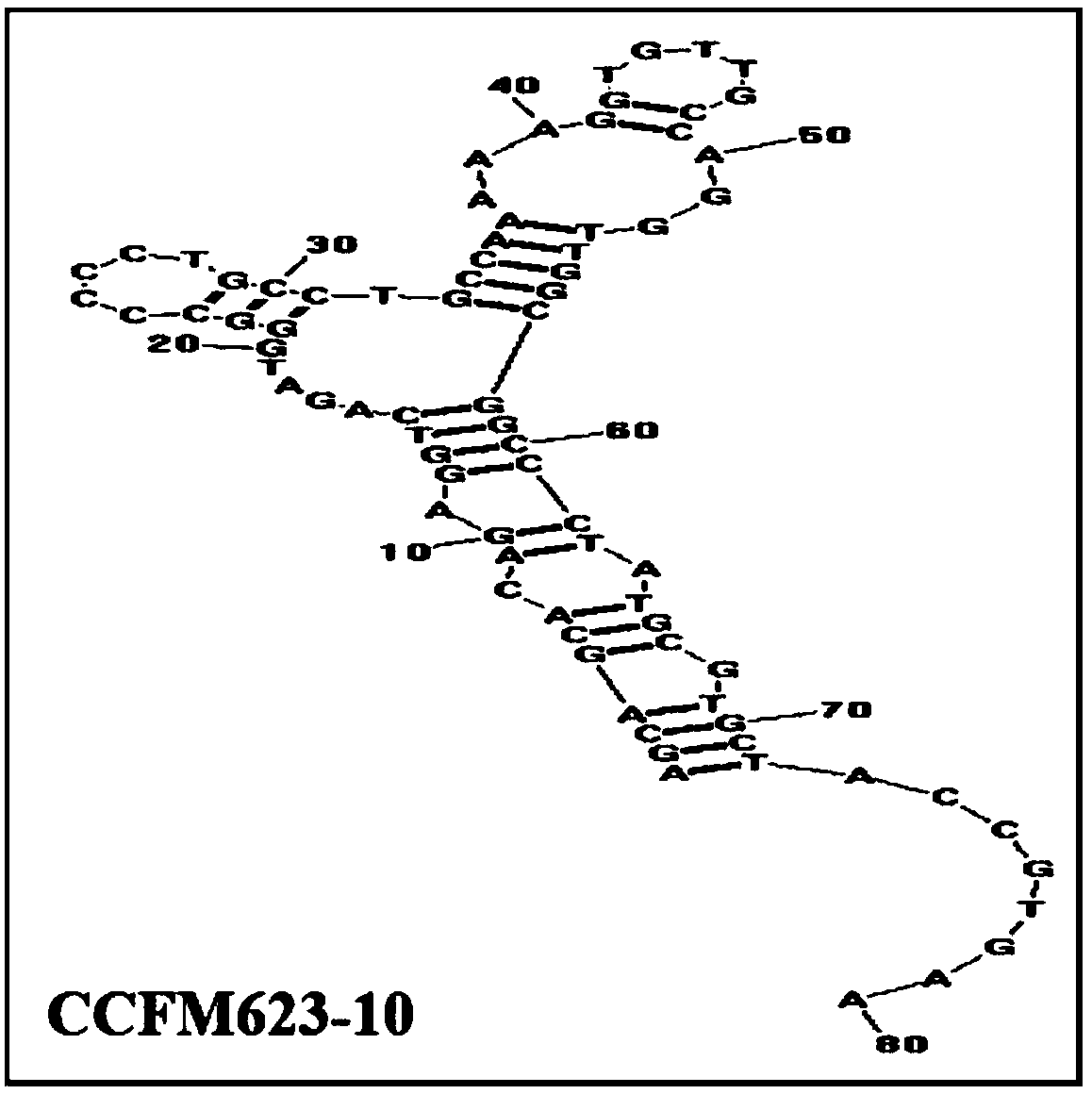
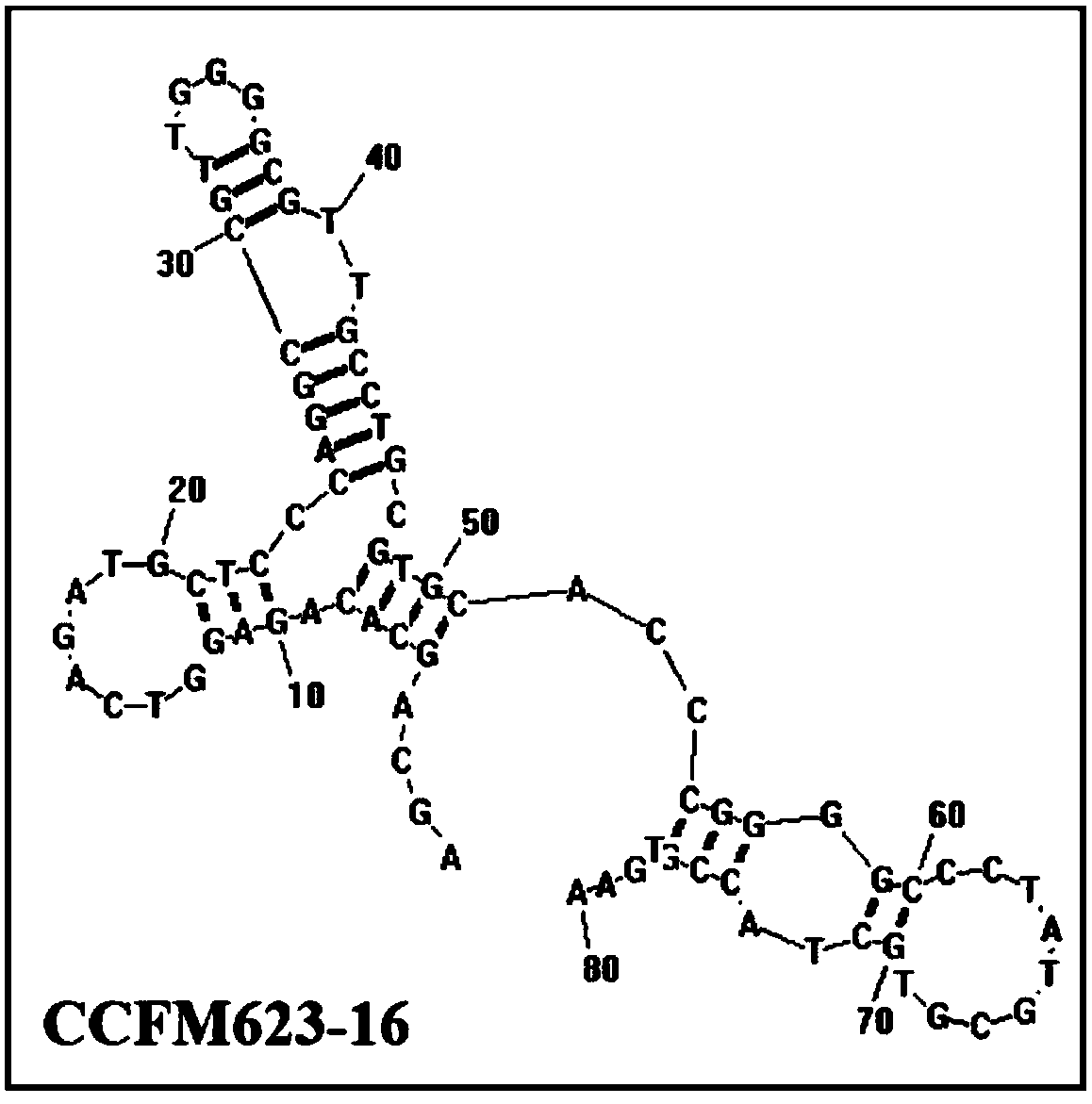
An aptamer, screening method and application for binding to Bifidobacterium breve
A technology of Bifidobacterium breve and aptamer, which is applied in the fields of biochemical equipment and methods, microbial determination/inspection, DNA preparation, etc. The method is simple, the preparation cycle is short, and the operation is convenient.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] (1) The initial random single-stranded DNA (ssDNA) library and primer library sequences were chemically synthesized in vitro as follows:
[0040] 5'-AGCAGCACAGAGGTCAGATG-N40-CCTATGCGTGCTACCGTGAA-3' (Integrated DNA Technologies, IDT), constructed a random ssDNA library with a length of 80 nt, with fixed primer sequences at both ends and a random sequence of 40 nt in the middle, with a library capacity of 10 14 above.
[0041]Upstream primer: 5'-AGCAGCACAGAGGTCAGATG-3'
[0042] Downstream primer: 5'-TTCACGGTAGCACGCATAGG-3'
[0043] 5' phosphorylation downstream primer: 5'-P-TTCACGGTAGCACGCATAGG-3'
[0044] The random ssDNA library and primers were prepared into 100uM stock solution with TE buffer and stored at -20°C for future use.
[0045] (2) SELEX screening
[0046] The Bifidobacterium breve that is cultivated to the logarithmic phase is used binding buffer (50mmol / L Tris-HCl (pH 7.4), 5mmol / L KCl, 100mmol / L NaCl, 1mmol / L MgCl 2 ) was washed twice to remove the cu...
Embodiment 2
[0048] The interaction between single-stranded nucleotide bases can make it form a certain spatial structure, and these structures are easy to realize conformational binding with various target molecules. Based on this conformational recognition feature, a sufficiently large capacity (10 14 The nucleic acid library of the above) can contain as many three-dimensional structures as possible, so it is possible to screen aptamers with high affinity and high specificity for any type of target.
[0049] Based on the above principles, the inventors in this example propose a set of nucleic acid aptamer sequences that specifically bind to Bifidobacterium breve through screening through improved whole-Bacterium SELEX technology. The present invention will be further described below in conjunction with the accompanying drawings and specific embodiments.
[0050] The nucleic acid aptamer screening method for the specific binding of Bifidobacterium breve is roughly as follows:
[0051] (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com