CrRNA that specifically recognizes gs gene and its application
A gene-specific technology, applied in the field of crRNA that specifically recognizes GS gene, can solve problems such as low efficiency and complex knockout process, and achieve the effects of high transfection efficiency, reduced clone number, and improved efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1: Construction of CRISPR-Cas9 expression system for GS gene
[0065] The GS gene of Chinese hamster ovary cells contains 7 exons, among which exons 2~7 encode the expression of GS protein ( figure 1 ), which plays a vital role in normal function. The crRNA sequences were designed for the 2-7 exons of GS gene. Using these crRNA sequences and Cas9 protein can make the expressed GS protein lose its function. The partial sequence of exon2~7 of GS is as follows (exons are marked with an underscore "_____"):
[0066] SEQ ID NO.17:
[0067] TGCCTCTTACGCAATTCCTGCAGGGGACCCCCTTCAGAGTAGATGTTAATGAAATGACTTTTGTCTCTCCAGAGCACCTTCCACC ATGGCCACCTCAGCAAGTTCCCACTTGAACAAAAACATCAAGCAAATGTACTTGTGCCTGC CCCAGGGTGAGAAAGTCCAAGCCATGTATATCTGGGTTGATGGTACTGGAGAAGGACTGCGCTGCAAAACCCGCACC CTGGACTGTGAGCCCAAGTGTGTAGAAG GTGAGCATGGGCAGGAGCAGGACATGTGCCTGGAAGTGGGCAAGCAGCCTGAGATTTGACCTTCCTTCTGTTTTGTTTGCAAAGTCTTTCAAAAGCAGGTCTCTTCAGGCCTCAGTCAGTCACCCGTAAGCTGCCGAGTAGTCTGGAGG
[0068] SEQ ID NO.18:
[0069] AC...
Embodiment 2
[0081] Example 2: Knockout and verification of GS gene on CHO-K1 cells
[0082] The designed crRNA and Cas9 protein expression vectors were transfected into CHO-K1 cells acclimated with CD medium, and a total of 16 transfections or combinations of A to P were designed.
[0083] CHO-K1 cells were purchased from ATCC, and DMEM / F12 basic medium was used for cell culture, supplemented with 5% fetal bovine serum. The expanded CHO-K1 cells were cultured in suspension using CD CHO medium, and glutamine and HT additives were added.
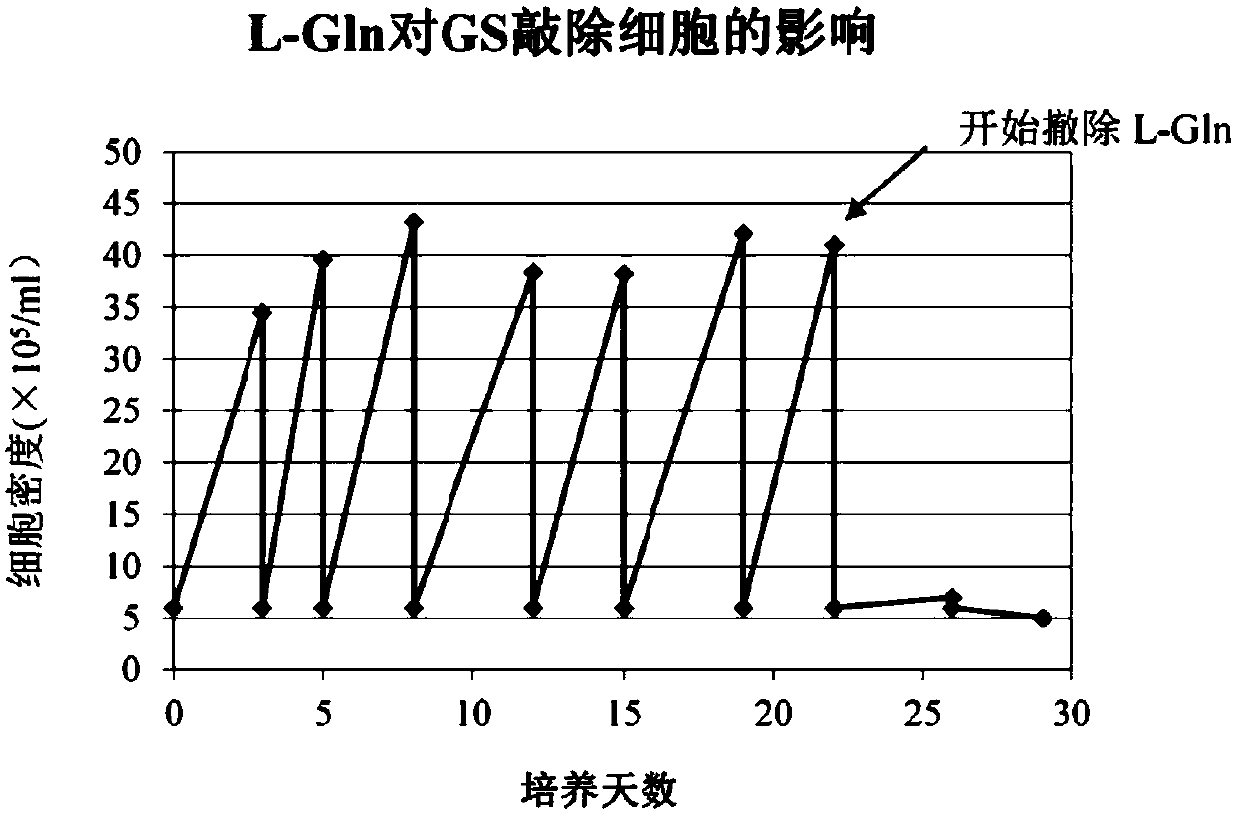
[0084] After the cells are adapted to suspension culture, the crRNA and Cas9 protein expression vectors are transfected into the cells. Puromycin was added to the cell culture medium after 48 hours of transfection. After 4 days of culture, the surviving cells were selected for subclonal culture. After the subclonal cells grow, the cells are transferred to L-Gln and L-Gln-free medium for culture. The clones that cannot grow in the medium without L-Gln but can ...
Embodiment 3
[0090] Example 3: Experiment of expressing antibodies in GS gene-deficient cells
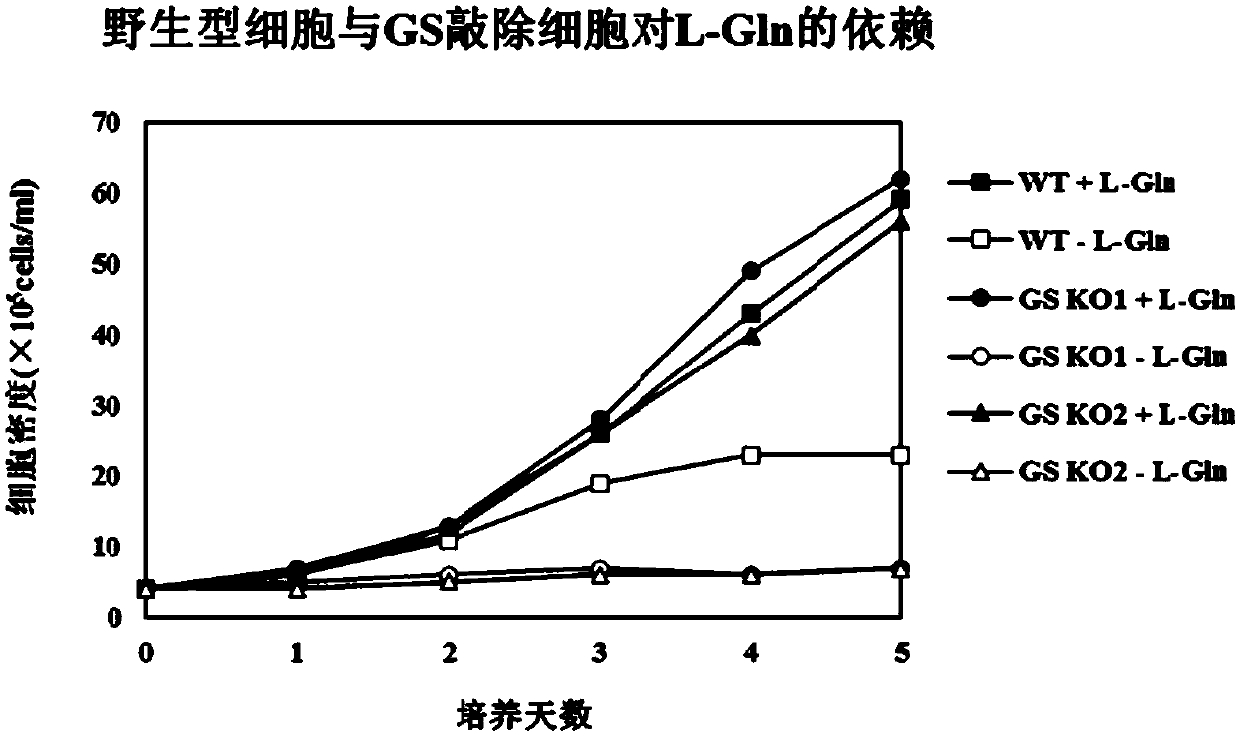
[0091] The selected GS KO1 and GS KO2 cells were subjected to antibody expression transfection experiments, and wild-type CHO-K1 cells were used as control hosts. KJ015 was transfected into GS KO1, GS KO2, CHO-K1 cells by exactly the same method. 48hrs after transfection, 25μM MSX was added as selection pressure. After 2 weeks of static culture, clones grew. The number of clones and the 25 clones with the highest antibody expression at 24hr were compared. The results are shown in Table 3. The differences in the number of clones grown from the three are relatively small, and the expression levels of GS KO1 and GS KO2 host clones are obviously higher.
[0092] table 3
[0093]
[0094]
[0095] The cells with better expression were expanded and cultured, and the expression products were collected, and the quality of expressed antibodies was analyzed using IEC as a key quality indicator. The results s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com