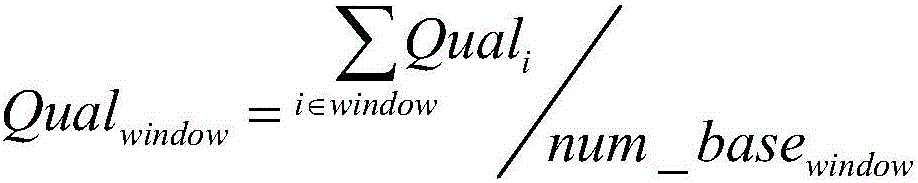Whole-exome sequencing data analysis system
A technology of sequencing data and whole exons, applied in digital data processing, special data processing applications, genomics, etc., can solve the problem of inability to identify low-frequency pathogenic variants and pathogenic mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0052] 1. Data introduction
[0053] Data Type: Whole Exome Sequencing
[0054] Tissue source: DNA from cancer tissue and peripheral blood of the same patient
[0055] Experimental Design: Exon Capture Sequencing
[0056] Sequencing platform: Illumina Hiseq 2000, paired-end sequencing
[0057] Average read length: 100bp
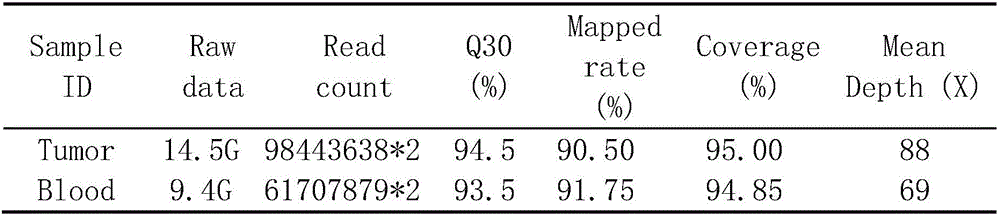
[0058] The quality statistics of the original sequencing data are shown in Table*.
[0059] Table 4.1 Whole-exome sequencing data quality statistics
[0060]
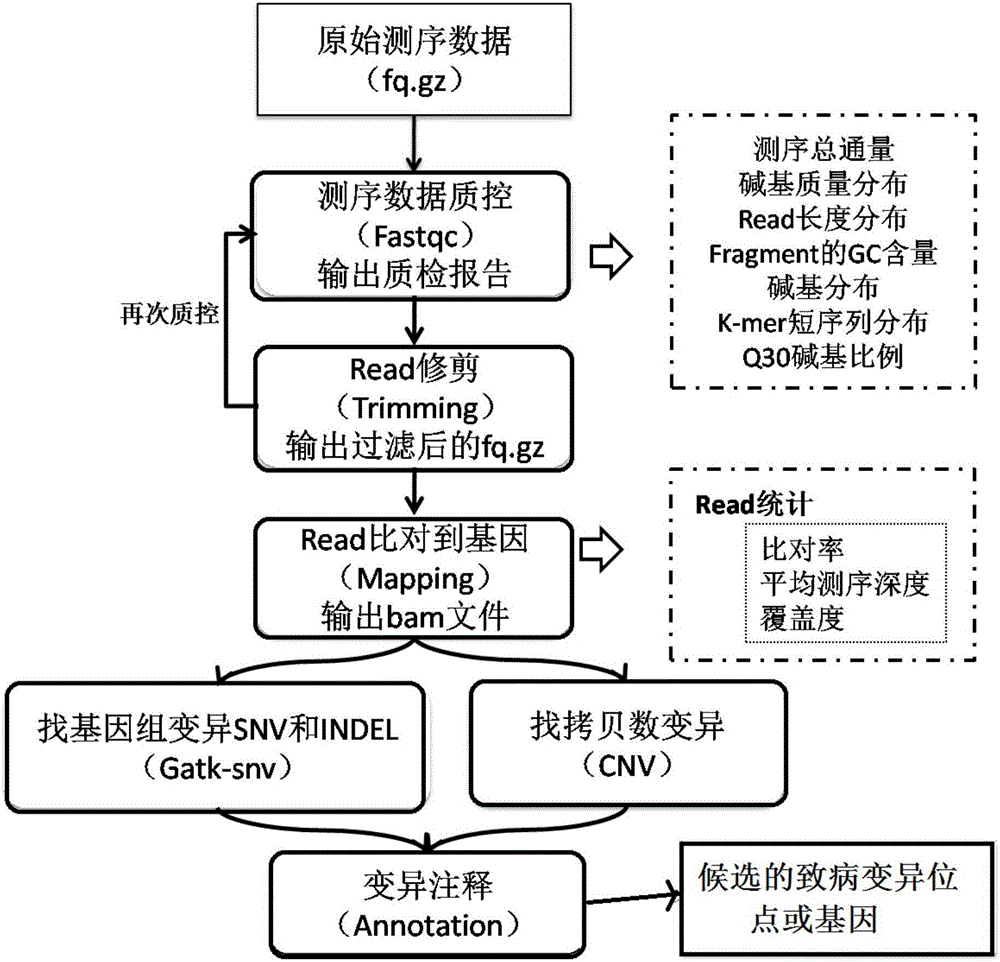
[0061] 2. System use
[0062] The whole exome sequencing data analysis process includes: sequencing data quality assessment and control, high-quality read screening, read comparison to the reference genome, searching for genomic variation, paired samples for somatic mutation, calculation of copy number variation, functional annotation, etc. . Next, each analysis step will be realized step by step by using the function modules integrated in the software.
[0063] (1) Quality control of raw se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com