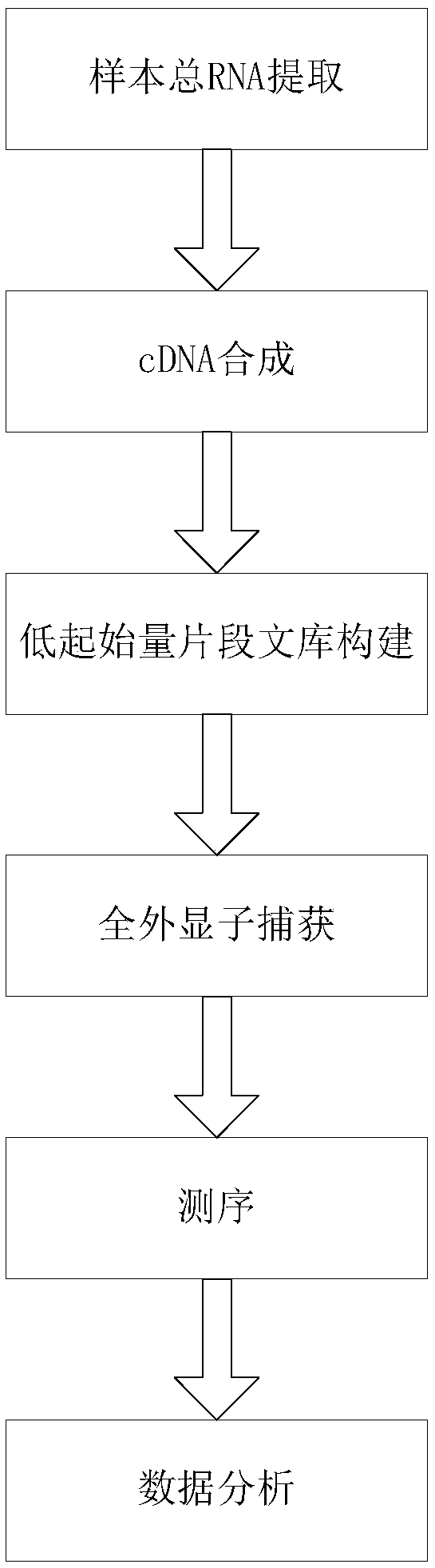
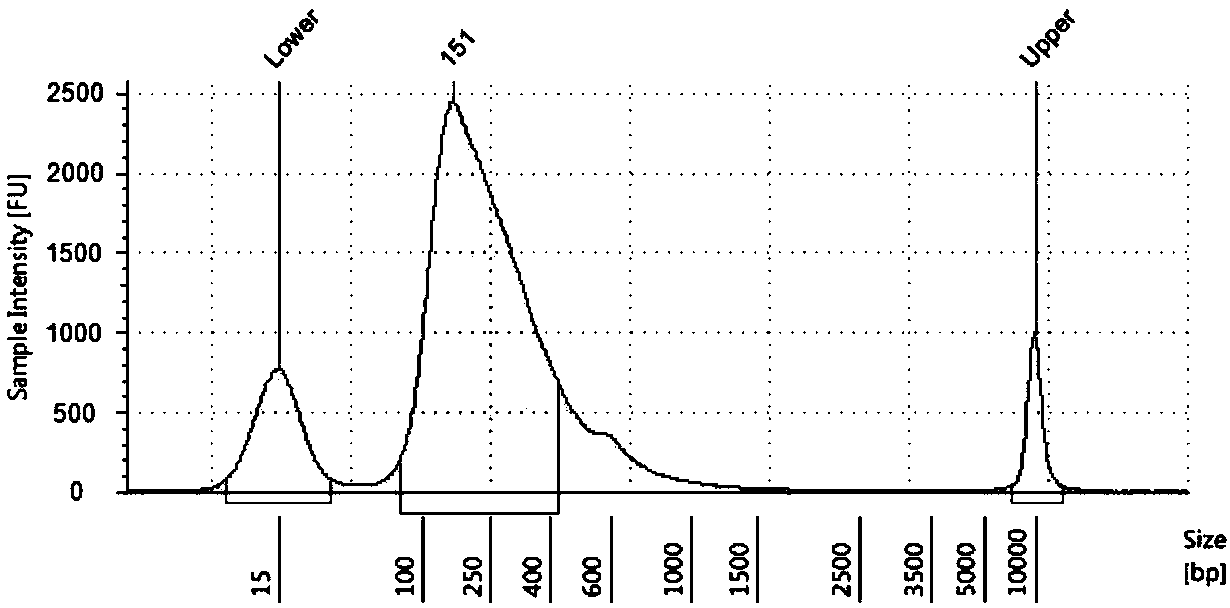
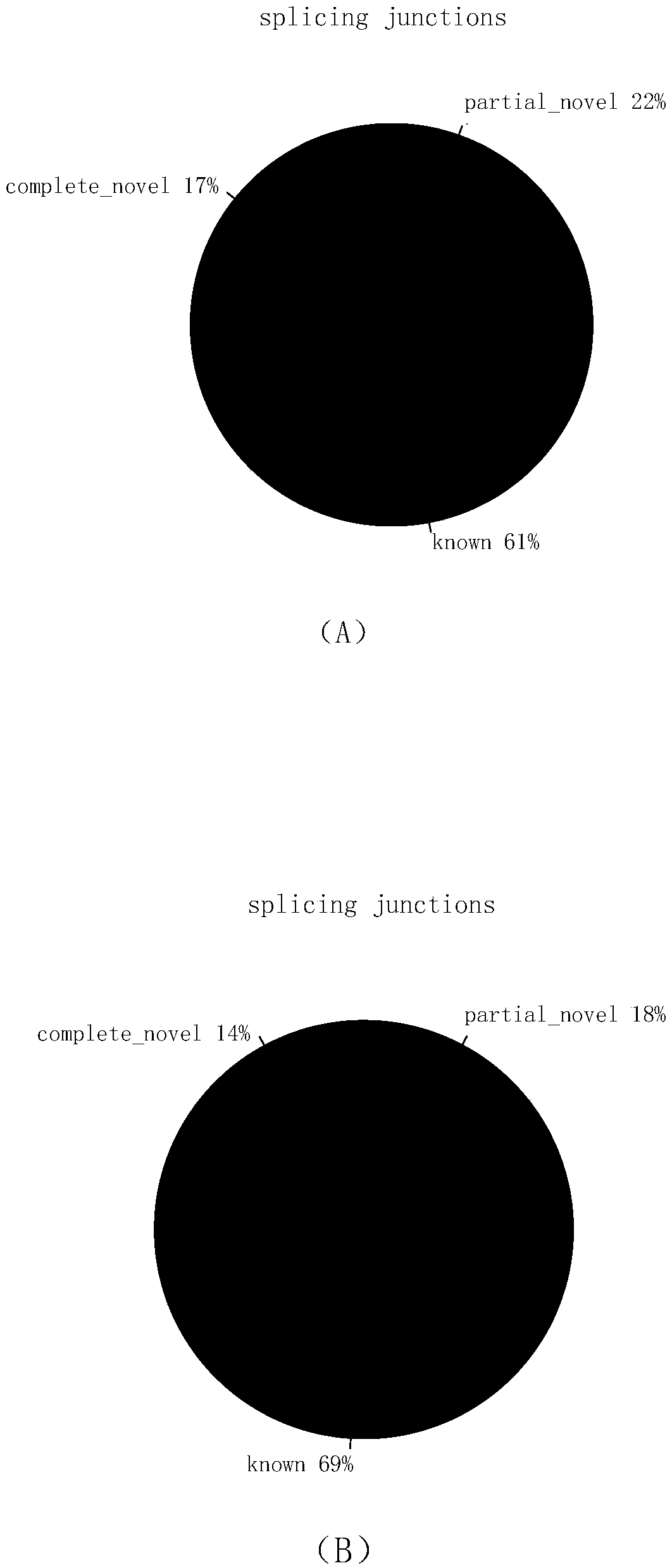Transcriptome sequencing method based on RT-WES (reverse transcription-whole exome sequencing) technology
A transcriptome sequencing and transcriptome technology, applied in the biological field, can solve the problems of inability to directly build a library and obtain valid data, high quality and input requirements, and difficulty in preserving fresh tissue, etc., to reduce further losses, low requirements, The effect of reducing quality and starting volume requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] 1. Extract the total RNA of the sample
[0033] Use MagMAX produced by Thermofisher TM FFPE DNA / RNA Ultra Kit (Cat. No. A31881) kit, according to the instructions of the kit, extract RNA from formalin-fixed and paraffin-embedded (Formalin-Fixed and Parrffin-Embedded, FFPE) tissue samples to obtain FFPE samples Total RNA.
[0034] 2. Synthesis of cDNA
[0035] 1. First strand synthesis
[0036] 1) Take a 0.2ml PCR reaction tube without nuclease contamination, add the ingredients listed in Table 1 (wherein, the random primer contains 6 bases), pipette and mix. This step needs to be operated in a safety cabinet that removes nuclease contamination, and ensure that the water used is free of nuclease contamination.
[0037] Table 1
[0038] ingredient
Volume (μl)
Total RNA of FFPE sample (100-500ng)
13.5
Random primer (50ng / μl)
1
[0039] 2) Place the above PCR reaction tube on a PCR machine, incubate at 65°C for 5 minutes, and quickly transfer it to an ice box after comple...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com