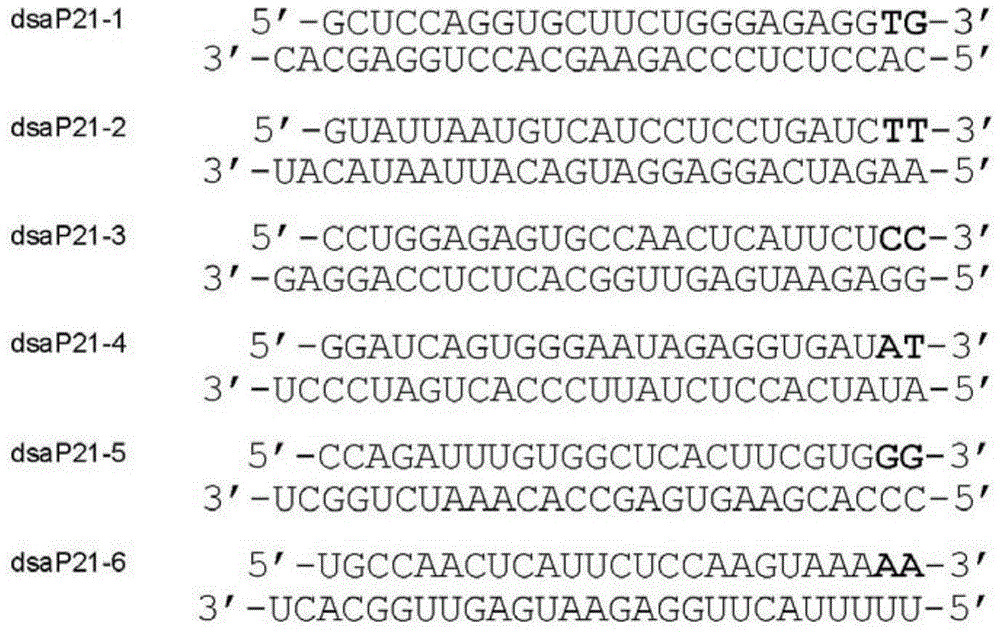Small-activating RNA, preparation method and applications thereof
A sense sequence and antisense sequence technology, applied in the application field of double-stranded small RNA in the field of RNA activation technology, can solve the problems of low saRNA design success rate and low RNA activation efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Materials and methods
[0052] 1. Design of Dicer substrate saRNA (dsaRNA):
[0053] The selection of the target site of the dsaRNA of the present invention is based on the following principles: 1. The selected target sequence is the sense sequence of the gene; 2. The 5' end of the guide RNA strand (guide RNA strand) is combined with the 3' end of the target sequence; 3. The GC content of the target sequence is 40-65%; 4. Avoid the target sequence containing 4 or more consecutive base sequences; 5. The 3' end of the target sequence is less thermodynamically stable than the 5' end. 6. The selected target sites should avoid CpG islands and high GC regions.
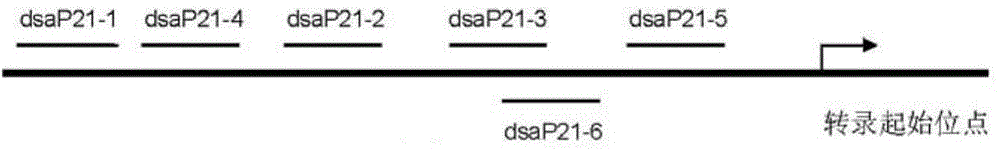
[0054] In the gene promoter region, the sequence length of the designed target site is 19 to 25 bases. The selected gene promoter region is from 5000 bases upstream of the transcription start site to the base before the gene transcription start site.
[0055] Taking the p21 gene as an example, the human p21 (CDKN1A...
Embodiment 2
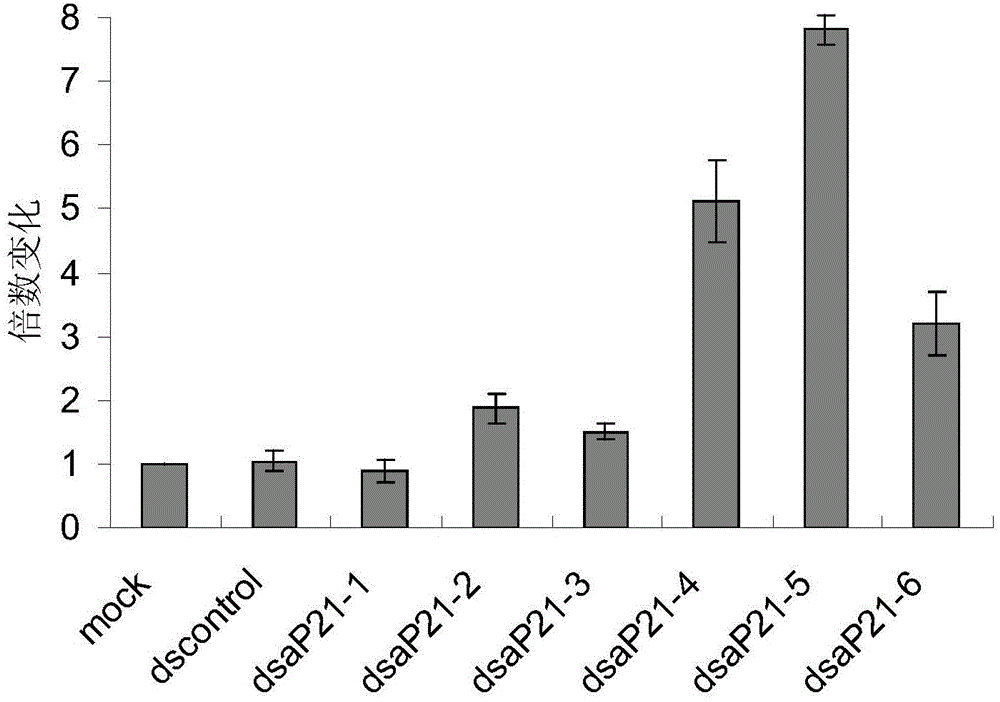
[0090] (1) Activation efficiency of saRNA shorter than 25 nucleotides
[0091]A dsRNA of 23 nucleotides in length was designed for the 6 sites of the p21 gene promoter. Named respectively: dsP21-1a, dsP21-2a, dsP21-3a, dsP21-4a, dsP21-5a, dsP21-6a. Its sequence is: dsP21-1a (sense sequence: SEQ ID NO: 30GCUCCAGGUGCUUCUGGGAGAGG, antisense sequence: SEQ ID NO: 31UCUCCCAGAAGCACCUGGAGCAC); dsP21-2a (sense sequence: SEQ ID NO: 32GUAUUAAUGUCAUCCCUCCUGATC, antisense sequence: SEQ ID NO dsP21-3a (sense sequence: SEQ ID NO: 34CCUGGAGAGUGCCAACUCAUUCU, antisense sequence: SEQ ID NO: 35AAUGAGUUGGCACUCUCCAGGAG); dsP21-4a (sense sequence: SEQ ID NO: 36GGAUCAGUGGGAAUAGAGGUGAT, antisense sequence: SEQ ID NO: 36GGAUCAGUGGGAAUAGAGGUGAT, antisense sequence: NO: 37CACCUCUAUUCCCACUGAUCCCT); dsP21-5a (sense sequence: SEQ ID NO: 38CCAGAUUUGUGGCUCACUUCGTG, antisense sequence: SEQ ID NO: 39CGAAGUGAGCCACAAAUCUGGCT); dsP21-6a (sense sequence: SEQ ID NO: 40UGCCAACUCAUUCUCCAAGUAAA, antisense sequence: S...
Embodiment 3
[0096] The dsaRNA designed according to the patent of the present invention can efficiently activate the expression of the pancreas-duodenum homeobox gene (PDX1). PDX1 gene is an important gene regulating islet function, and PDX1 can also promote the expression of insulin gene in non-β cells such as liver cells, which has important application value for the treatment of diabetes. We designed dsaRNA targeting different sites in the PDX1 gene promoter region. The deoxyribonucleotides in the dsaRNA sequence are indicated in lower case bold in the sequences shown below. dsaPDX1-1 (sense sequence is: SEQ ID NO: 54CACACUAUGUCCAUUAUCAAAUA ta, antisense sequence is: SEQ ID NO: 55UAUAUUUGAUAAUGGACAUAGUGUGUU); dsaPDX1-2 (sense sequence is: SEQ ID NO: 56CCGACAUCUUUGUGGCUGUGAACaa, antisense sequence is: SEQ ID NO: 57UUGUUCACAGCCACAAAGAUGUCGGUU); dsaPDX1-3 (sense sequence is: SEQ ID NO: 58GACCUAGAGAGCUGGGUCUGCAAac, antisense sequence is: SEQ ID NO: 59GUUUGCAGACCCAGCUCUCUAGGUCAG); dsaPDX1-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com