High-flux chip processing and analysis flow control method for long-chain non-coding RNA
A long-chain non-coding, process control technology, applied in the direction of electrical digital data processing, special data processing applications, instruments, etc., can solve the problems of long-chain non-coding RNA high-throughput chip data processing inaccuracy, incomprehensible analysis, etc. , to achieve the effect of optimizing working time, wide application range and simple realization method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
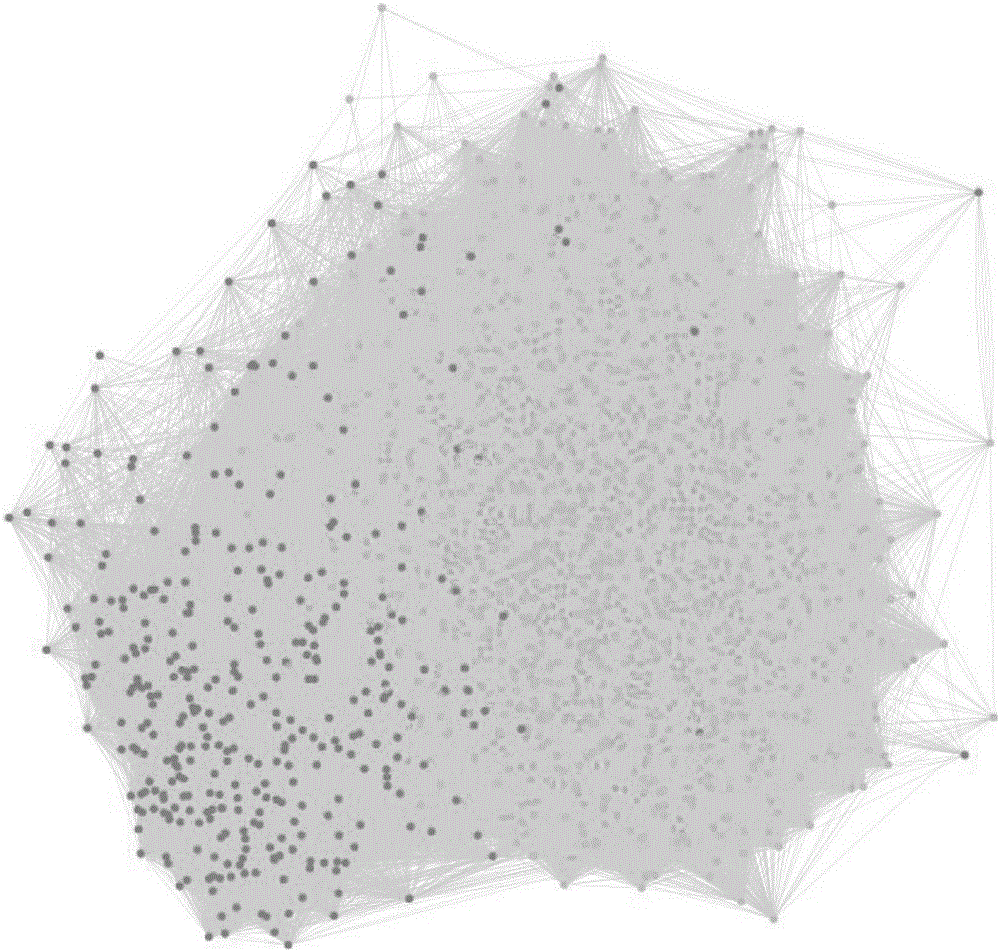
[0097] Firstly, the original data is filtered, and then the low-quality signal and noise-contaminated data are removed, and the effective long-chain non-coding RNA expression value is obtained after normalization. Based on the analysis results of long-chain non-coding RNA, the target gene can be predicted based on its sequence characteristics; the function prediction can also be combined with the co-expressed gene expression profile. Long noncoding RNA-coexpressed gene networks such as image 3 shown. On the basis of the above analysis, a series of statistical and visual analysis can be carried out.
[0098] 1. The original signal file of long non-coding RNA is shown in Table 1
[0099] Analysis platform: R platform
[0100] Analysis software: Affy, limma
[0101] Table 1
[0102]
[0103] Explanation of column names:
[0104]
[0105] 2. The expression results of the long-chain non-coding RNA chip are shown in Table 2
[0106] Analysis platform: R platform
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com