Recombinant herpes simplex virus HSV-hTERTp_ICP4_Hep-GFP and corresponding diagnostic kit thereof
A herpes simplex virus, virus genome technology, applied in the direction of virus, virus/phage, double-stranded DNA virus, etc., can solve the problem of low virus reproduction titer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] A preparation method for recombinant herpes simplex virus HSV-hTERTp_ICP4_Hep-GFP, comprising the following steps:
[0085] 1) Replace the ICP4 gene promoter in the herpes simplex virus containing the ICP4 gene with the human telomerase reverse transcriptase promoter hTERTp to construct the recombinant herpes simplex virus HSV-hTERTp_ICP4:
[0086](1) Construction of shuttle plasmids pICP4del-hTERTp_ICP4 and pICP4del-eGFP
[0087] a. Cultivate the herpes simplex virus containing ICP4 gene with BHK cells, and purify its genomic DNA;
[0088] b. Amplify the flanking sequence upstream of the ICP4 gene: with the viral genome DNA obtained in step a) as a template, use the following ICP4USf forward primer and ICP4USr reverse primer:
[0089] ICP4USf forward primer: ccctccagacgcaccggagtcggggg,
[0090] ICP4USr reverse primer: aagtcgactctagaggatcgatctctgacctgagattggcggcactgaggta
[0091] Amplify the upstream flanking sequence of the ICP4 gene;
[0092] c. Amplify the downst...
Embodiment 2
[0143] Embodiment 2HSV_Hep-GFP selectively propagates and produces high-titer virus in liver cancer cells:
[0144] HSV_HEP-GFP was used to infect liver cancer cells (HuH7, HepG-2, 7721), lung cancer cells (A549, PG), gastric cancer cells BGC823, colon cancer cells HT-29, human erythroleukemia cells TF-1, lymph Tumor cell U937, breast cancer cell MD-MB-231, pharyngeal squamous cell carcinoma cell Fadu, and melanoma cell A375 were tested for virus titer at 6, 12, 24, 48, and 72 hours. The results are shown in Table 1. Experimental data show that HSV_HEP-GFP has the best effect on infecting liver cancer cells, and the virus reproduction titer after 24 hours or later is significantly higher (more than 2 logarithms) than that of other cancer cells that can reproduce the virus.
[0145]
Embodiment 3
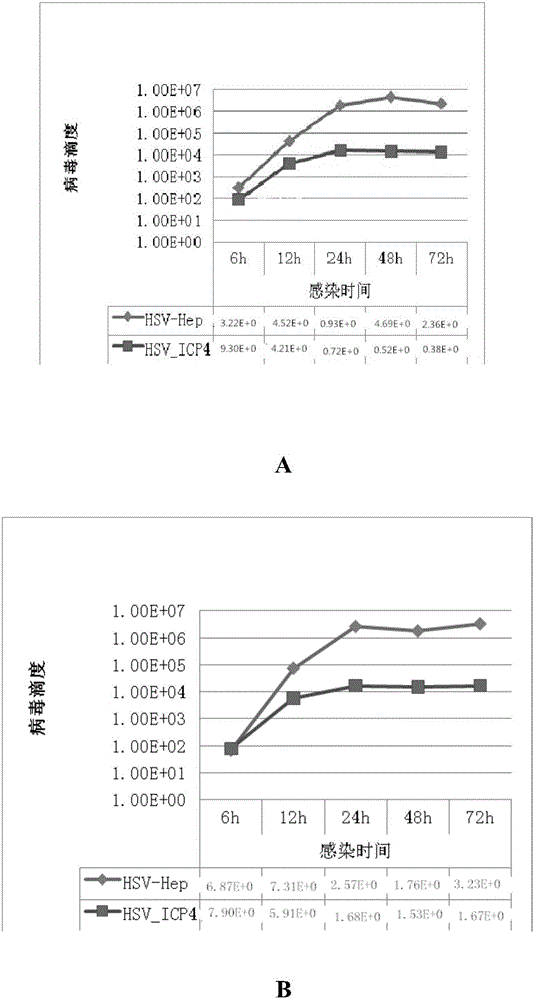
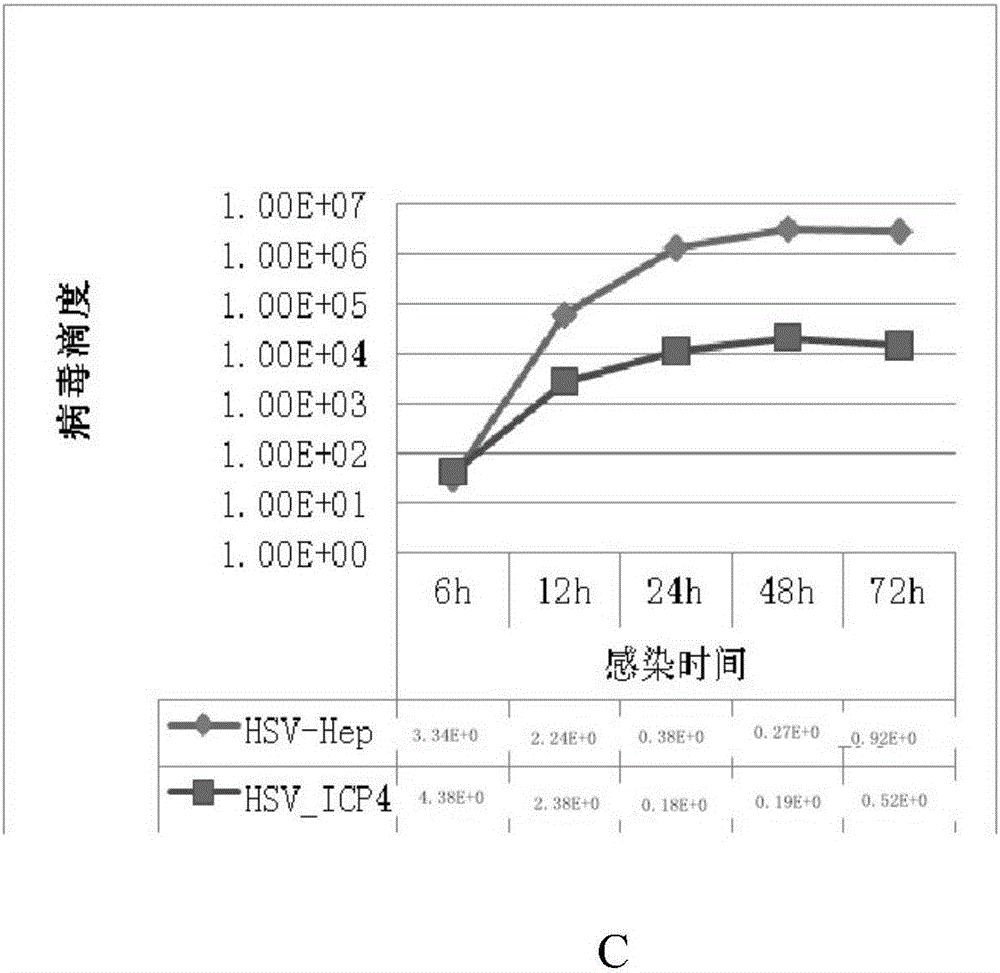
[0146] Embodiment 3HSV_Hep-GFP produces the virus of higher titer than HSV_ICP4 in liver cancer cell
[0147] HSV_HEP and HSV_ICP4 were respectively infected with liver cancer cells HuH7, HepG-2, and 7721 at MOI=0.01, and the virus titers were detected at 6, 12, 24, 48, and 72 hours. see results figure 1 -3. Experimental data showed that HSV_HEP infected three kinds of liver cancer cells, and the viral reproduction titers reached 10 after 24 hours. 6 Above, although HSV_ICP4 can reproduce in three kinds of liver cancer cells, but the virus titer is low, the peak value is only 10 4 . The two viruses infected liver cancer cells, and their reproductive titers differed by 2 logarithms.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com