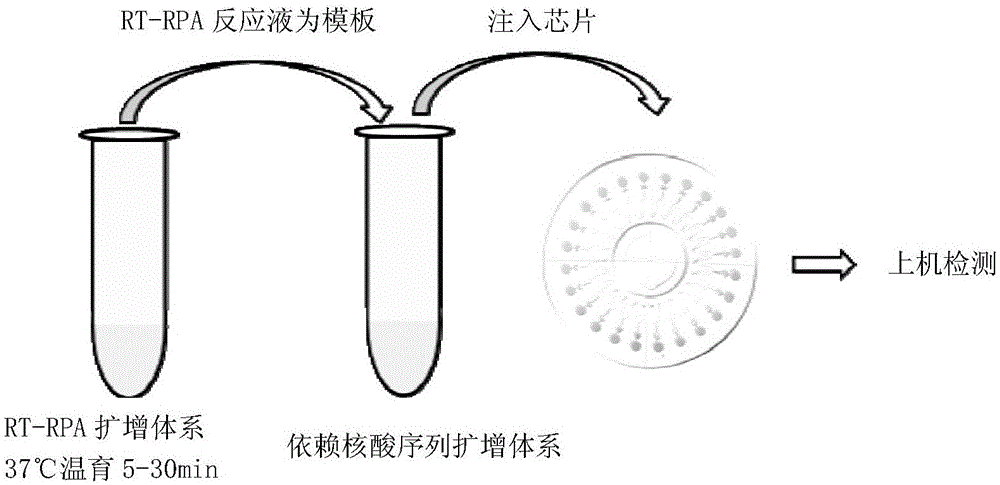
Nucleic acid amplification technology and application thereof in detection of mosquito-borne viruses
A nucleotide, dengue virus technology for recombinant DNA technology, resistance to vector-borne diseases, microorganism-based methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0141] Embodiment 1, design and prepare each primer and probe
[0142] The primer pair used to detect Zika Virus (Zika Virus) consists of the following two primers (5'→3'):
[0143] ZIKV-F (Sequence 1 of the Sequence Listing): TGCATWGGAGTCAGCAA;
[0144] ZIKV-R (SEQ ID NO: 2 of the SEQUENCE LISTING): AATTCTAATACGACTCACTATAGGGAGATAGGATCTTACTCVGCCA.
[0145] The primer pair used to detect Chikungunya virus (Chikungunya virus) consists of the following two primers (5'→3'):
[0146] CHIKV-F (Sequence 3 of the Sequence Listing): CCGACTCAACCATCCTGGAT;
[0147] CHIKV-R (SEQ ID NO: 4 of the Sequence Listing): AATTCTAATACGACTCACTATAGGGAGAGGCARACGCAGTGGTACTTCCT.
[0148] The primer pair used to detect dengue virus type Ⅰ consists of the following two primers (5'→3'):
[0149] DENV1-F (Sequence 5 of the Sequence Listing): CAAAAGGAAGTCGTGCAATA;
[0150] DENV1-R (SEQ ID NO: 6 of the Sequence Listing): AATTCTAATACGACTCACTATAGGGAGACTGAGTGAATTCTCCTACTGAACC.
[0151] The primer pair used...
Embodiment 2
[0179] Embodiment 2, the preparation of mosquito-borne virus detection kit
[0180] 1. The composition of the kit
[0181] The kit consists of the following components (each component is individually packaged):
[0182] (1) The primer mixture consists of ZIKV-F, ZIKV-R, CHIKV-F, CHIKV-R, DENV1-F, DENV1-R, DENV2-F, DENV2-R, DENV3-F, DENV3-R, DENV4-F , DENV4-R and sterile water; in the primer mixture, the concentration of ZIKV-F is 0.2 μM, the concentration of ZIKV-R is 0.2 μM, the concentration of CHIKV-F is 0.15 μM, and the concentration of CHIKV-R is 0.15 μM , DENV1-F at a concentration of 0.2 μM, DENV1-R at a concentration of 0.2 μM, DENV2-F at a concentration of 0.15 μM, DENV2-R at a concentration of 0.15 μM, DENV3-F at a concentration of 0.3 μM, DENV3-R The concentration was 0.3 μM, the concentration of DENV4-F was 0.25 μM, and the concentration of DENV4-R was 0.25 μM.
[0183] (2) RT-RPA reaction buffer: TwistDx company, component of TABARTS01kit.
[0184] (3) Reactio...
Embodiment 3
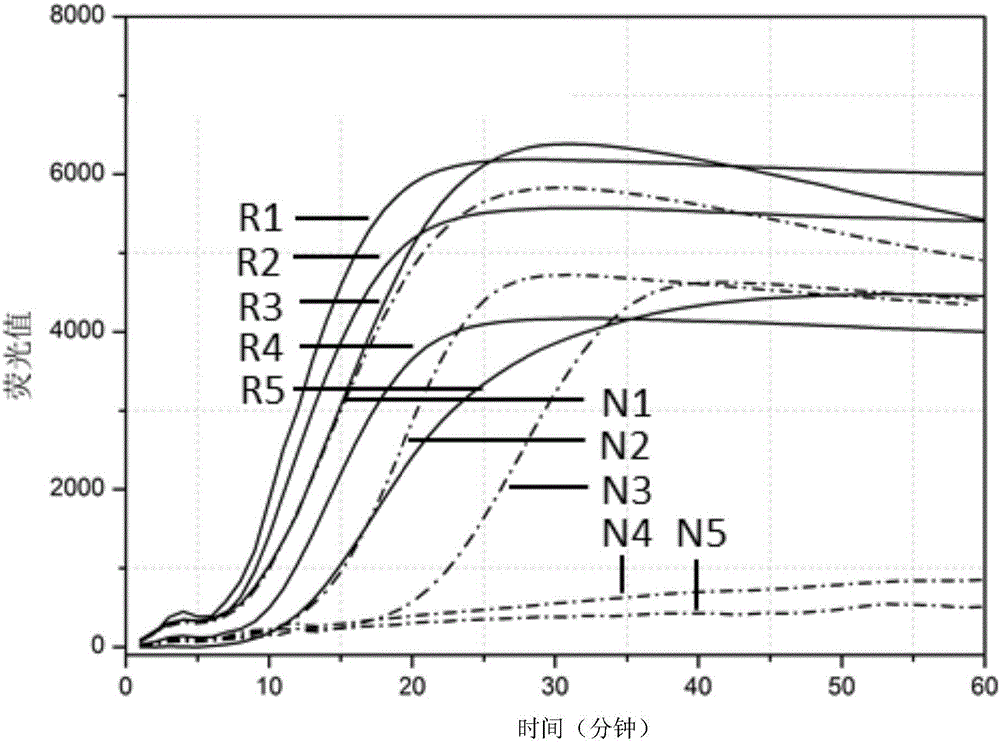
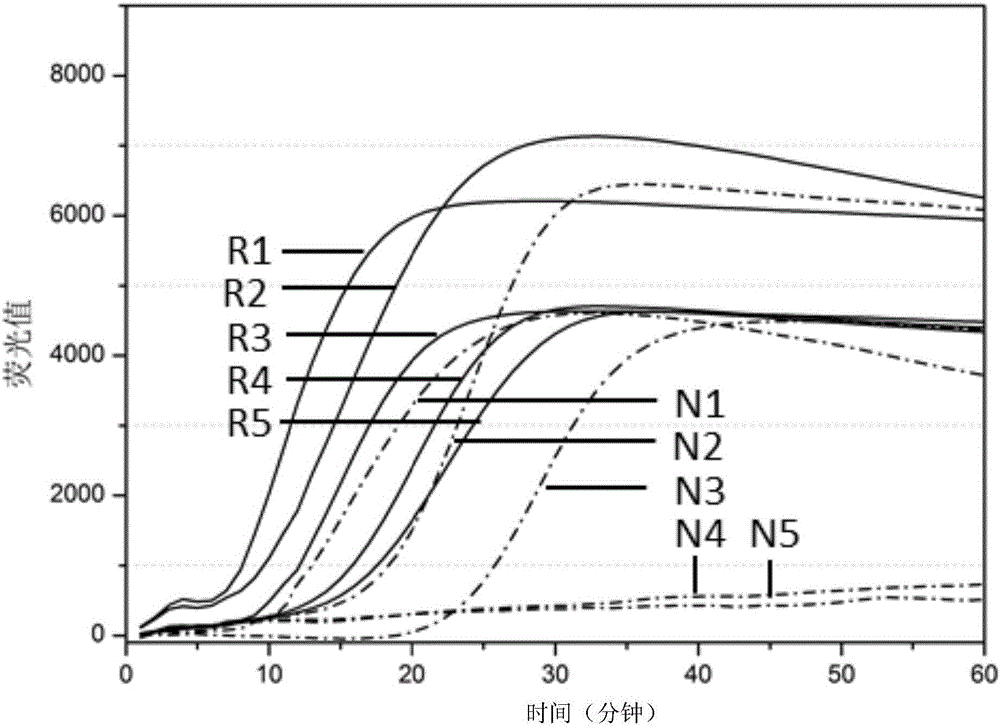
[0206] Embodiment 3, the contrast of two kinds of nucleic acid amplification methods
[0207] 1. Preparation of standard products
[0208] RNA1 (single-stranded RNA molecule, Zika virus standard) shown in sequence 22 of the sequence listing was prepared. Dissolve RNA1 in sterile water, and then perform serial dilution with sterile water to obtain a concentration of 10 4 copies / μl, 10 3 copies / μl, 10 2 copies / μl, 10 copies / μl or 1 copies / μl of RNA1 dilution.
[0209] RNA2 (single-stranded RNA molecule, chikungunya virus standard) shown in sequence 23 of the sequence listing was prepared. Dissolve RNA2 in sterile water, and then perform serial dilution with sterile water to obtain a concentration of 10 4 copies / μl, 10 3 copies / μl, 10 2 copies / μl, 10 copies / μl or 1 copies / μl of RNA2 dilution.
[0210] RNA3 (single-stranded RNA molecule, dengue virus type I standard) shown in sequence 24 of the sequence listing was prepared. Dissolve RNA3 in sterile water, and then perfor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com