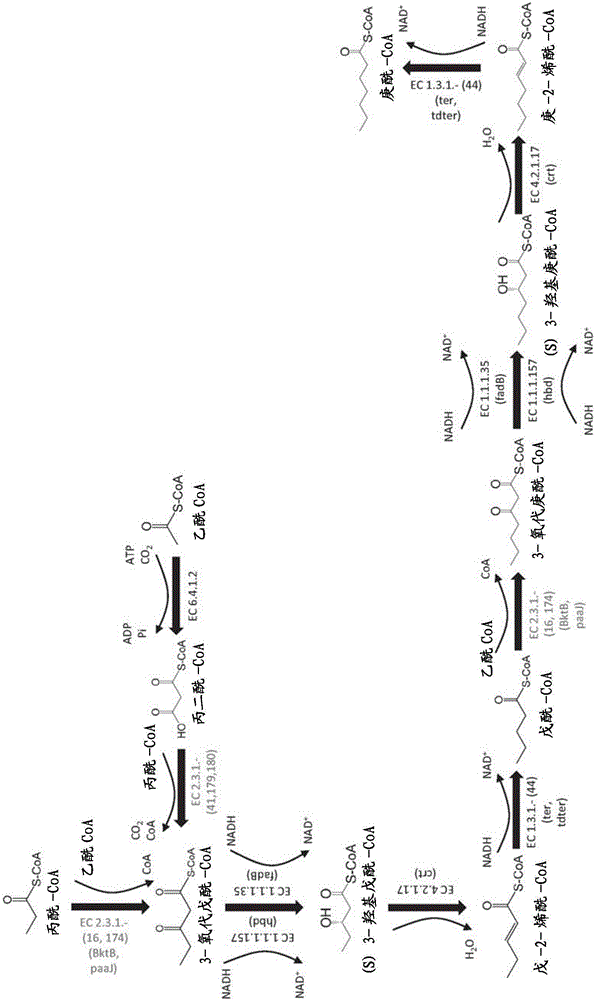
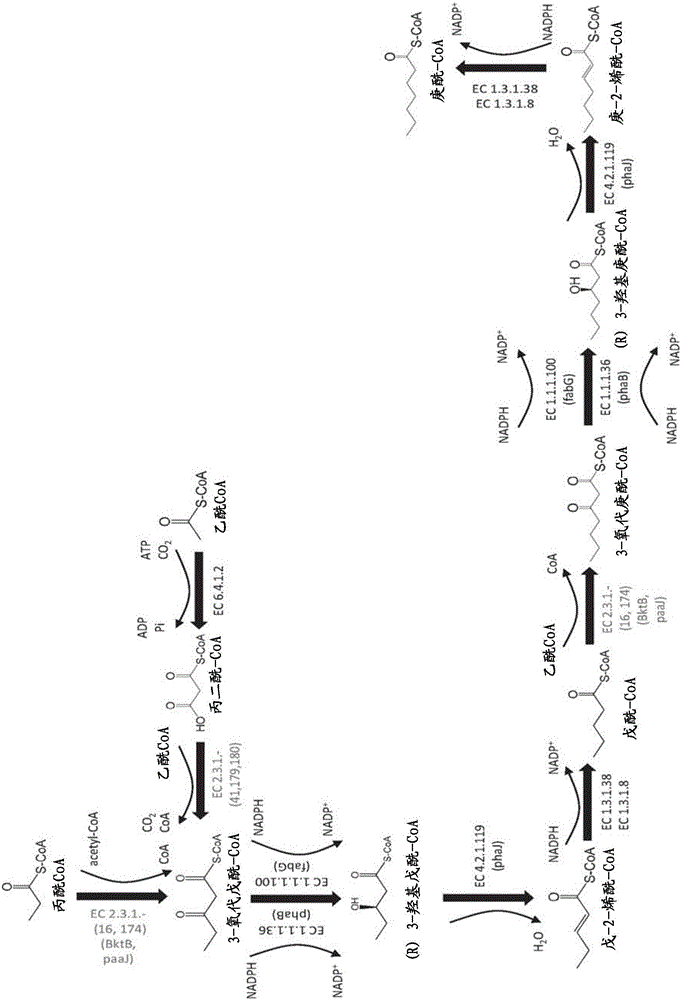
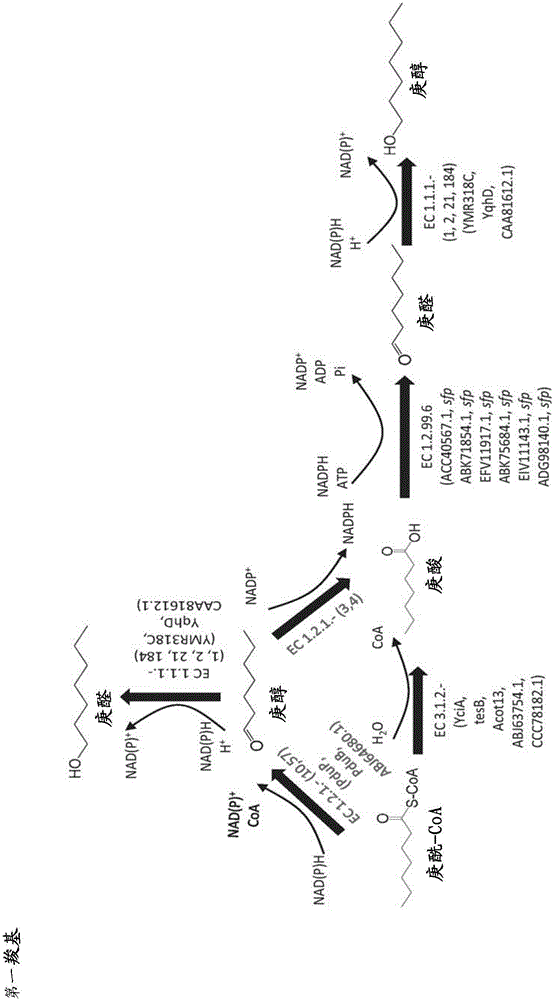
Methods, reagents and cells for biosynthesizing compounds
A reactive, hydroxyalkyl-based technology for biosynthesis of compounds, reagents and cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0373] Enzymatic activity of omega-transaminase using pimelate semialdehyde as substrate and forming 7-aminoheptanoic acid
[0374] The nucleotide sequence encoding the N-terminal His-tag was added to the genes from Chromobacter violaceum, Pseudomonas syringae, Rhodobacter sphaericus and Vibrio fluvialus encoding the ω-transaminases of SEQ ID NO: 8, 11 and 13, respectively (see Figure 10), allowing the generation of N-terminally HIS-tagged ω-transaminases. Each of the resulting modified genes was cloned into the pET21a expression vector under the control of the T7 promoter, and each expression vector was transformed into a BL21[DE3] E. coli host. The resulting recombinant E. coli strains were grown in 250 mL shake flask cultures containing 50 mL LB medium and antibiotic selection pressure at 37° C. with shaking at 230 rpm. Each culture was induced overnight at 16°C with 1 mM IPTG.
[0375] The pellet from each induced shake flask culture was harvested by centrifugation. E...
Embodiment 2
[0381] Carboxylate reductase activity using pimelic acid as a substrate and forming pimelic semialdehyde
[0382] The nucleotide sequence encoding the His-tag was added to the genes from Segniliparus rugosus and Segniliparus rotundus encoding the carboxylic acid reductases of SEQ ID NOs: 4 (EFV11917.1) and 7 (ADG98140.1), respectively (see Figure 10 ), allowing the generation of an N-terminally HIS-tagged carboxylic acid reductase. Each modified gene was cloned into the pET Duet expression vector together with the sfp gene encoding the His-tagged phosphopantetheinyltransferase from Bacillus subtilis, both under the T7 promoter. Each expression vector was transformed into a BL21[DE3] E. coli host, and each resulting recombinant E. coli was grown in a 250 mL shake flask culture containing 50 mL LB medium and antibiotic selection pressure at 37°C with shaking at 230 rpm strain. Each culture was induced overnight at 37°C using autoinduction medium.
[0383] The pellet from eac...
Embodiment 3
[0387] Enzyme activity of carboxylic acid reductase using 7-hydroxyheptanoic acid as substrate and forming 7-hydroxyheptanal
[0388] The nucleotide sequence encoding the His-tag was added to the carboxylic acid reductases from Mycobacterium marinum, Mycobacterium smegmatis, Segniliparus rugosus, Mycobacterium smegmatis, Marseilles respectively encoding the carboxylic acid reductase of SEQ ID NO:2-7. Mycobacterium, and Segniliparus rotundus genes (GenBank accession numbers ACC40567.1, ABK71854.1, EFV11917.1, ABK75684.1, EIV11143.1, and ADG98140.1, respectively) (see Figure 10 ), allowing the generation of an N-terminally HIS-tagged carboxylic acid reductase. Each modified gene was cloned into the pET Duet expression vector together with the sfp gene encoding the His-tagged phosphopantetheinyl transferase from Bacillus subtilis, all under the control of the T7 promoter. Together with the expression vector from Example 3, each expression vector was transformed into a BL21[DE3]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com