Parallel probe, gene chip, kit and assay method for clostridium difficile assay
A technology for detecting gene chips and detection kits, applied in the field of molecular biology, can solve the problems of insufficient sensitivity, short detection period, low specificity, etc., and achieve the effects of improving accuracy and precision, rapid detection, and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 Design of primers and probes for target fragments
[0023] Find the gene sequence of Clostridium difficile in the NCBI database, select the highly specific gene sequence as the target sequence (ID number: NC_009089.1), and design primers and probes. After the target sequence is determined, according to the primer and probe design principles, design the amplification primers and probes of the C. difficile target gene. The 5'end of the R primer of the C. difficile target gene amplification primer is labeled with Cy3. The specific sequence is as follows :
[0024] C-DIFF target sequence amplification primers:
[0025] F primer (C-DIFF-F): 5'-gagagtttgatyctggctcag-3' (SEQ ID NO. 8);
[0026] R primer (C-DIFF-R): 5'-Cy3-aaggaggtgatccarccgca-3' (SEQ ID NO. 9).
[0027] C-DIFF parallel probe:
[0028] C-DIFF probe 1:
[0029] 5’-ttttttttttttttttcagcaacgccgcgtgagtgatgaaggccttcgggtcgtaa-3’ (SEQ IDNO.1)
[0030] C-DIFF probe 2:
[0031] 5’-ttttttttttttttttaccagttgcgaaggcggctctctgga...
Embodiment 2
[0042] Example 2 Preparation of chip
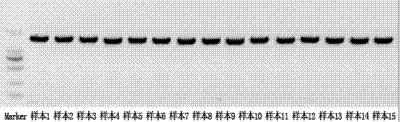
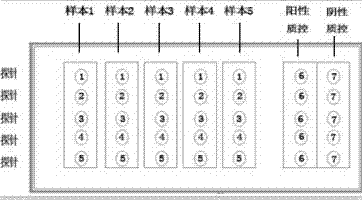
[0043] Spot the parallel probes of C-DIFF in Example 1 on the amino-modified glass substrate in accordance with the order in Table 1, to obtain a gene chip containing probes (such as figure 1 Shown). The probe concentration is 30uM, each spot is 0.2uL, and then incubated at 80°C for 1.5 hours.
[0044] Table 1 The sequence of the probes of the Clostridium difficile detection chip
[0045]
Embodiment 3
[0046] Example 3 Detection method
[0047] 1. Extract the genome of the sample to be tested
[0048] Use the viral genomic DNA / RNA co-extraction kit to extract the genome of the sample to be tested as the amplification template according to the steps in the operating instructions.
[0049] 2. PCR amplification of the target fragment
[0050] After a lot of experiments, the PCR amplification system and PCR reaction program of the target gene detected by C-DIFF were determined. The PCR amplification system with a total volume of 10 μL contains the following reagents:
[0051]
[0052] The PCR reaction program is: 95°C 5min; 95°C 10s, 55°C 30s, 72°C 30s, 30 cycles; 72°C 10min.
[0053] 3. Hybridization of PCR product and chip
[0054] Follow the steps below: a. On the gene chip prepared in Example 2, spot the PCR amplification products of the sample to be tested into the detection holes of 5 probes, and at the same time, the positive control sample and the blank control sample The PCR prod...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com