Method for detecting miRNA-155 based on aptamer
A nucleic acid aptamer, mir-155 technology, applied in the field of biotechnology detection, can solve the problems of difficulty in satisfying miR-155 convenient, fast, high-sensitivity detection, complex sample preprocessing, low detection sensitivity, etc., and achieves fast detection speed. , low detection limit, simple preparation method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
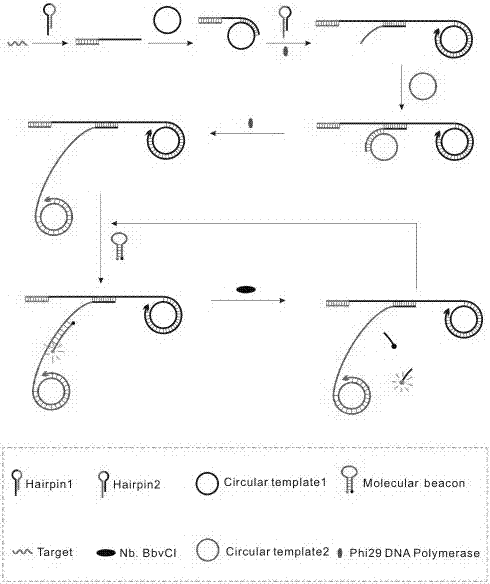
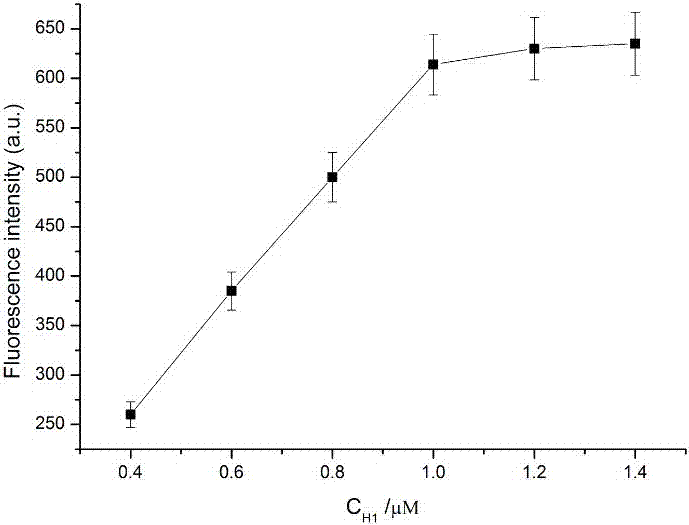
[0042]a. Mix 1 μL of different concentrations of Hairpin1 (final concentrations are 0.4 μM, 0.6 μM, 0.8 μM, 1.0 μM, 1.2 μM, 1.4 μM), Circular template1 (1 μL, 10 μM), Hairpin2 (1 μL, 10 μM), Circular template 2 (1μL, 10μM), Molecular beacon (1μL, 100μM), phi29 DNA polymerase (1μL, 10×), Nb.BbvCI (1μL, 10×), NEB buffer (5μL, 10×) and the test substance (miRNA-155) were added to the EP tube, shaken for 30s, and placed in a water bath at 37°C for 70min.
[0043] b. Take the reacted solution out of the water bath, put it into a water bath at 85°C and heat it in a water bath for 10 minutes to inactivate the enzyme in the system.
[0044] c. After 10 min, take out the mixed solution from the water bath. Then an Agilent fluorescence detector was used to measure the fluorescence intensity of the mixed solution, and the target object was detected accordingly.
[0045] Its experimental principle diagram is as figure 1 , see the result figure 2 , it can be seen from the figure tha...
Embodiment 2
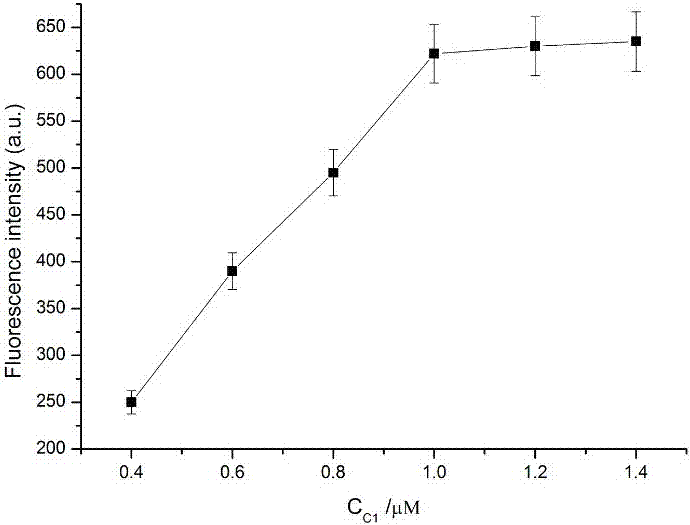
[0047] a. Mix 1 μL of different concentrations of Circular template1 (final concentrations are 0.4 μM, 0.6 μM, 0.8 μM, 1.0 μM, 1.2 μM, 1.4 μM), Hairpin1 (1 μL, 10 μM), Hairpin 2 (1 μL, 10 μM), Circular template 2 (1μL, 10μM), Molecular beacon (1μL, 100μM), phi29 DNA polymerase (1μL, 10×), Nb.BbvCI (1μL, 10×), NEB buffer (5μL, 10×) and the test substance (miRNA-155) were added to the EP tube, shaken for 30s, and placed in a water bath at 37°C for 70min.
[0048] b. Take the reacted solution out of the water bath, put it into a water bath at 85°C and heat it in a water bath for 10 minutes to inactivate the enzyme in the system.
[0049] c. After 10 min, take out the mixed solution from the water bath. Then an Agilent fluorescence detector was used to measure the fluorescence intensity of the mixed solution, and the target object was detected accordingly.
[0050] see results image 3 , it can be seen from the figure that the detected fluorescence intensity value increases as...
Embodiment 3
[0052] a. Add 1 μL of different concentrations of Hairpin2 (0.4 μM, 0.6 μM, 0.8 μM, 1.0 μM, 1.2 μM, 1.4 μM), Hairpin1 (1 μL, 10 μM), Circular template 1 (1 μL, 10 μM), Circular template 2 (1μL, 10μM), Molecular beacon (1μL, 100μM), phi29 DNA polymerase (1μL, 10×), Nb.BbvCI (1μL, 10×), NEB buffer (5μL, 10×) and the test substance (miRNA-155) were added to the EP tube, shaken for 30s, and placed in a water bath at 37°C for 70min.
[0053] b. Take the reacted solution out of the water bath, put it into a water bath at 85°C and heat it in a water bath for 10 minutes to inactivate the enzyme in the system.
[0054] c. After 10 min, take out the mixed solution from the water bath. Then an Agilent fluorescence detector was used to measure the fluorescence intensity of the mixed solution, and the target object was detected accordingly.
[0055] see results Figure 4 , it can be seen from the figure that the detected fluorescence intensity value increases as the concentration of Ha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com