Applications of Xanthomonas pathopoiesis-associated gene
A Xanthomonas and gene technology, applied in the application field of genes in plant disease prevention and control, can solve the problems of crop yield reduction, harvest failure, economic loss, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
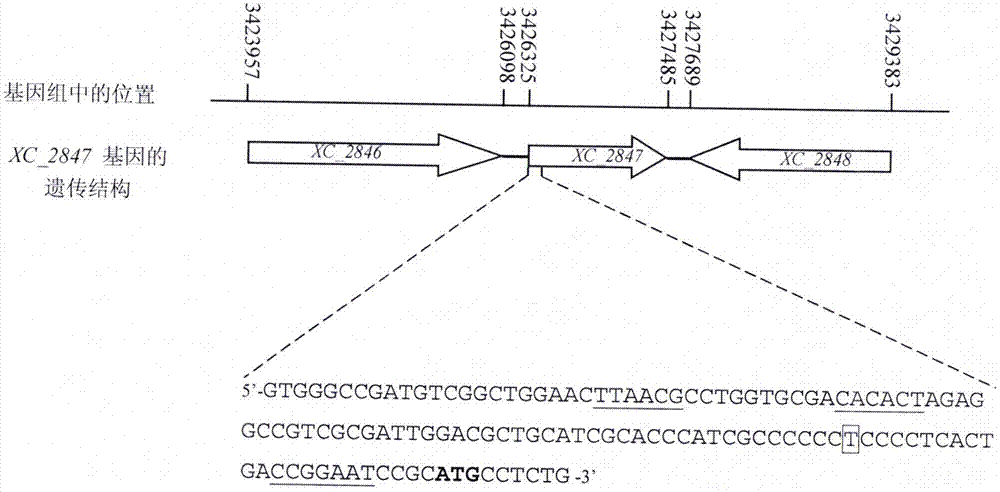
[0063] Determination of the transcription initiation site of the gene of Brassicaceae black rot fungus XC_2847 (hpaM, hereinafter referred to as XC_2847 gene)
[0064] Since the XC_2847 gene was annotated as encoding a "hypothetical protein" in the whole genome sequencing of the Xcc 8004 strain, whether this gene is a gene with a definite function is the first question to be determined. Here, first use 5'-RACE (i.e. 5' end rapid amplification method, an experimental method in molecular biology) to determine the transcription start site of XC_2847 gene, the specific method can refer to the description of literature Li Ruifang et al. (Li Ruifang et al. et al., 2014, please refer to the references listed below for details).
[0065] For this, 4 nested primers 2847RTP1-4 need to be designed:
[0066] 2847RTP1: AGATAGATGCCCTGGCTGCC;
[0067] 2847 RTP2 CGGTGCGATTGAACGACACG;
[0068] 2847RTP3: CGGTGCTGGCCACGATCTGCAT;
[0069] 2847RTP4: ATCGGCCATGCGCAGCACGG).
[0070] Culture Xcc...
Embodiment 2
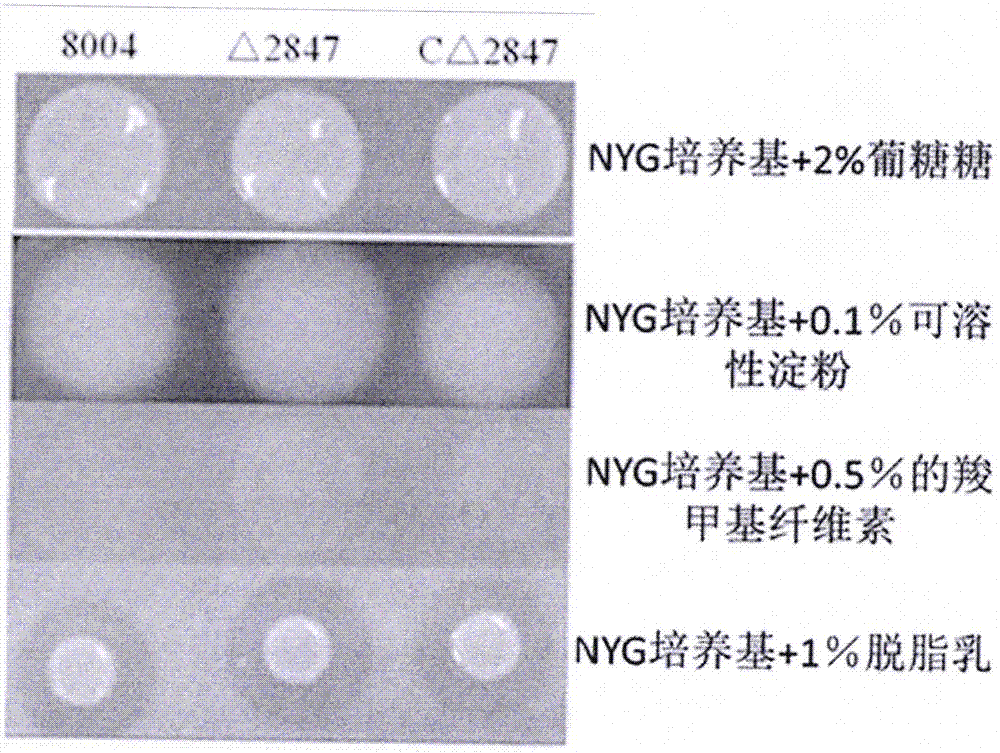
[0077] Construction of XC_2847 gene mutant of Brassicaceae black rot fungus
[0078] To construct a deletion mutant of XC_2847, refer to the specific method The method described by et al. (Gene 1994, 145:69-73).
[0079] According to the DNA sequence of the upstream and downstream of the XC_2847 gene (the genome sequence number of the Xcc 8004 strain is: NC_007086), design primers:
[0080] 2847UF: ACAGTT GAATTC GATGGCGTCAACCAGCTCTC;
[0081] 2847UR: ACAGTT TCTAGA TCTAGTGTGTCGCACCAGGC)
[0082] 2847DF: ACAGTT TCTAGA GATGTGGCCGGCGTGGATATC;
[0083] 2847DR: ACAGTT AAGCTT ATTCCTGGACGTGGCGATGC;
[0084] Using the total DNA of Brassicaceae black rot fungus 8004 strain as a template, the PCR method (pre-denaturation at 95°C for 4 minutes; denaturation at 95°C for 1 minute, renaturation at 55°C for 30 seconds, extension at 74°C for 1 minute, 30 cycles; extension at 74°C for 5 minutes) The DNA homologous fragments of 747bp upstream and 726bp downstream of the XC_2847...
Embodiment 3
[0087] Cloning and sequence determination of XC_2847 gene and functional complementation of XC_2847 gene mutant
[0088] According to the DNA sequence of XC_2847 gene (NC_007086), design primers
[0089] 2847F: ACAGTT GGATCC GCCTACAGCTTCTGAGCCTG;
[0090] 2847R: ACAGTT AAGCTT GTGGCTGCGTAGCGGTTTTG;
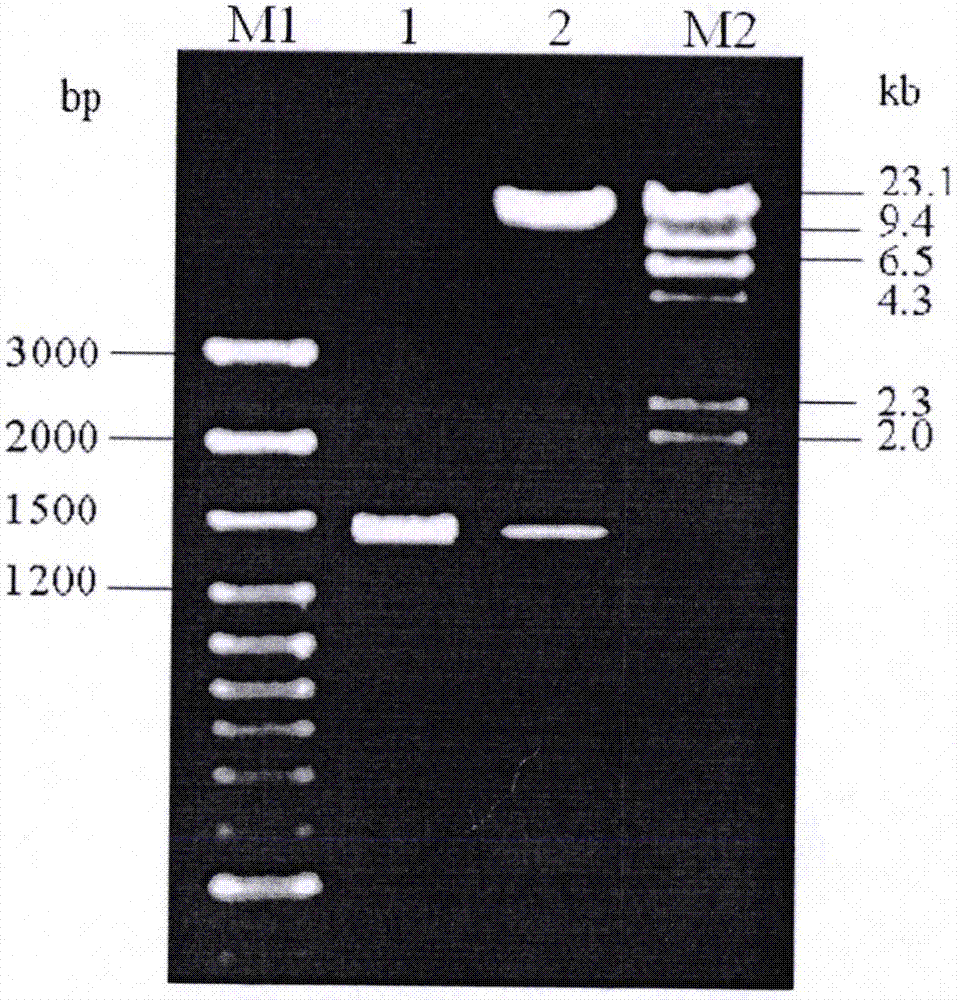
[0091] Using the total DNA of the Xcc 8004 strain as a template, PCR method (pre-denaturation at 95°C for 4 min; denaturation at 95°C for 1 min, annealing at 55°C for 30 s, extension at 74°C for 2 min, and 30 cycles; extension at 74°C for 5 min) was used to amplify the entire DNA containing the XC_2847 gene. A long 1432bp DNA sequence. For the convenience of cloning, the restriction site sequences of BamHI and HindIII (that is, the underlined part of the above DNA sequence) were added to the 5' ends of the primers.
[0092] The obtained DNA fragment was cloned into the vector pGEM3Zf(+), and the DNA nucleotide sequence was determined on an ABI377 DNA automatic sequencer by...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com