High-throughput sequencing-based detection kit for SNP loci and detection method for detection kit
A detection kit and detection method technology, applied in the field of forensic genetics, can solve the problems of limited detection efficiency, unable to meet the needs of forensic individual identification and kinship identification, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
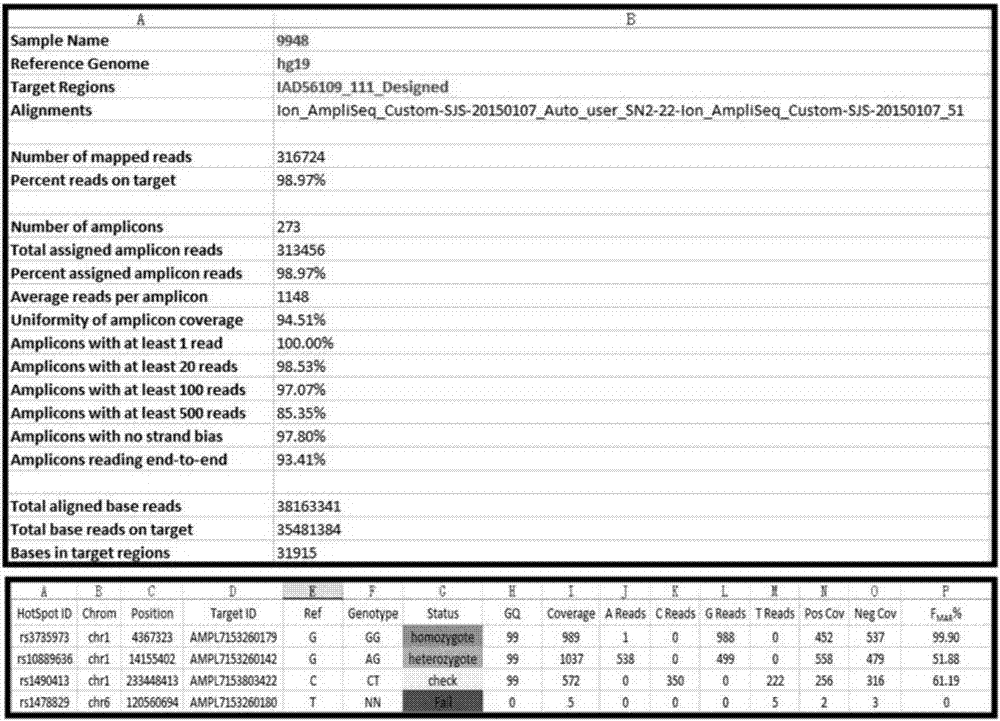
[0065] This example is the typing result of the standard product DNA9948.
[0066] Its detection method is as follows:
[0067] Step 1) Take 1 μL standard product DNA9948 (10ng / μL) for library construction, the PCR system is 20 μL, which contains 5 μL 2.5×PCR reaction buffer II, 2 μL 5× primer mixture, 1 μL Taq polymerase (2U), 1 μL standard Product DNA 9948 and 11 μL deionized water. PCR conditions for library construction: 95°C pre-denaturation for 11 minutes; 94°C for 30s, 60°C for 60s, 70°C for 60s, 22 cycles; 60°C for 30min.
[0068] Step 2) purifying and quantifying the library constructed in step 1). Purification is done using Ampure magnetic beads, and quantification can be done by Qubit or qPCR. This time, qPCR was used to complete the library quantification.
[0069] Step 3) Ligate adapters and sequencing primers to the library quantified in step 2).
[0070] Step 4) Sequence the library prepared in step 3) on a high-throughput sequencing platform (Ion Torrent f...
Embodiment 2
[0081] This example is the application of the SNP site detection kit of the present invention in the identification of suspected mutations in triplet relatives.
[0082] A case of triplet kinship testing involving the biological mother-child (female)-suspect father. The detection method described in Example 1 was adopted for operation.
[0083] STR detection results based on CE technology: 19 autosomal STR loci were detected using the conventional PCR-CE detection kit Goldeye 20A (Beijing Basic Cognitive Biology Co., Ltd.), and it was found that the mother and child were completely identical at the 19 STR loci. According to Mendel's law of inheritance, the suspect father and child had one-step mutations at two STR loci (vWA and FGA). Through calculation, the cumulative parentage index CPI is 378.361. According to the triplet paternity test standard, it is impossible to judge the parent-child relationship between the suspicious father and the child. Supplementary detection Th...
Embodiment 3
[0087] This example is the forensic detection efficiency of the SNP site detection kit of the present invention in 50 unrelated individuals.
[0088] The blood samples of 50 unrelated individuals were tested using the detection method described in Example 1. 50 individuals were volunteers.
[0089] result:
[0090] (1) 50 individuals were completely typed at 234 autosomal SNP loci and 30 X-SNP loci, and 25 male individuals were completely genotyped at 9 Y-SNP loci.
[0091] (2) According to the frequency data of 50 unrelated individuals at 273 SNP sites, it was found that 234 autosomal SNP sites were independent of each other, and the cumulative individual identification rate for individual identification was 1-1.49E-62, which was used for doublet The cumulative non-paternal exclusion rate for body paternity testing is 1-2.98E-12, and the cumulative non-paternal exclusion rate for triplet paternity testing is 1-3.51E-23, which can meet the actual work needs. The detection d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com