Method and device for determining pre-rearrangement V/J gene sequences
A technology for identifying devices and J genes, applied in the field of bioinformatics, can solve problems such as difficulty in accurate assembly and correction, and influence on germline inference, and achieve the effect of reducing difficulty, shortening time and cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
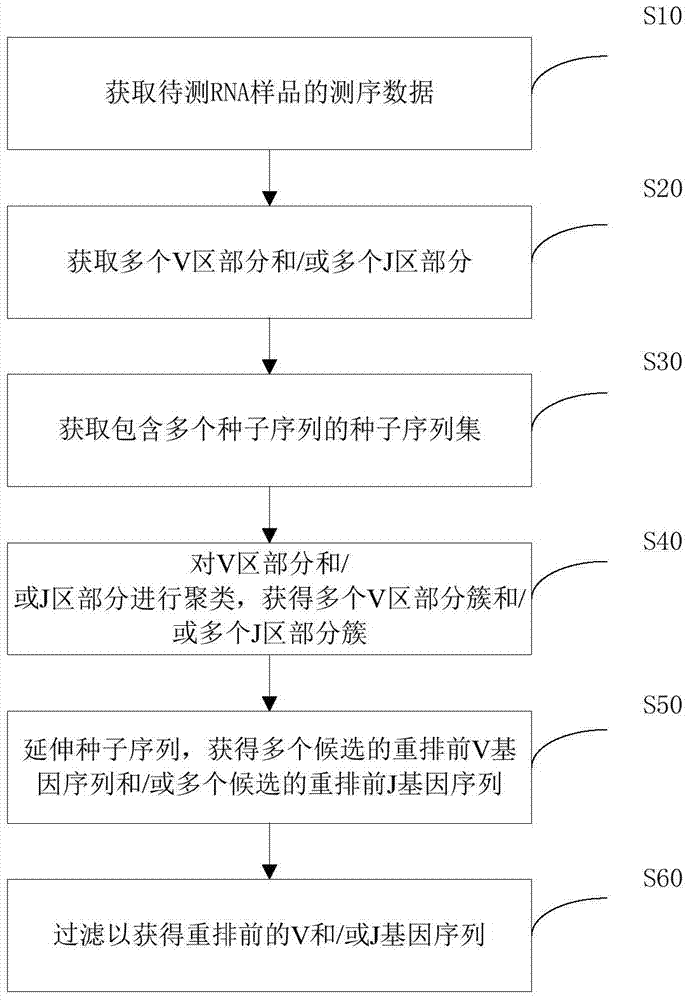
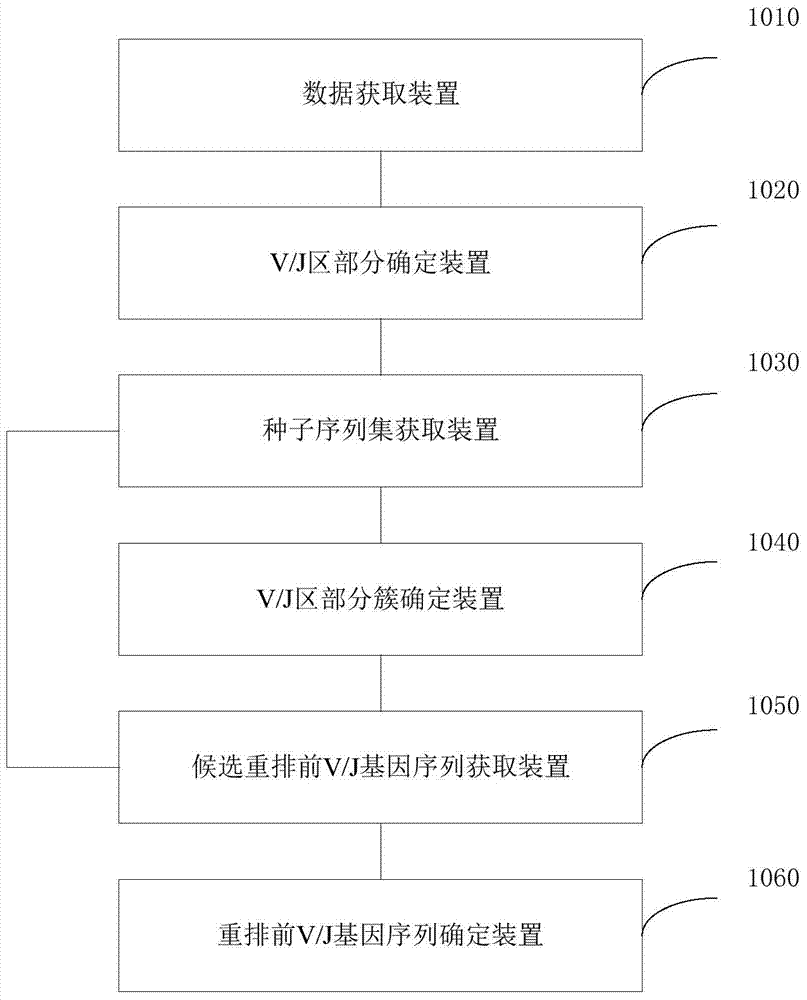
[0070] The general method includes the following steps:
[0071] For RNA samples, a set of universal primers optimized by the inventors can be used to amplify the variable region of TCR, BCR or Ig by 5'race:
[0072]The variable region is formed by the rearrangement of the V, D, and J gene segments of TCR or Ig. During the rearrangement process, there are nucleotide insertions and deletions at the junctions between the gene segments. This region reflects the surface of adaptive immune molecules. Diversity of receptors. The C region is a constant region. For RNA, primers can be designed in the C region to amplify the variable region, and then the variable region obtained by the rearrangement of the V region and J region of different subfamilies can be amplified by the 5'race method.
[0073] (2) Library preparation
[0074] Step 1 Synthesize the first strand of cDNA through the reverse transcription primer of the C region and Superscript II, and then digest the RNA in the cDN...
Embodiment 2
[0097] (1) Experimental process
[0098] (1) 5'RACE enrichment of the target fragment
[0099] The peripheral blood of three normal people was drawn, and the peripheral blood mononuclear cells (peripheral blood mononuclear cell PBMC) were separated to extract RNA, and three RNA samples were obtained, which were recorded as sample 1 (HRB), sample 2 (HXY) and sample 3 (XHS). RNA is reverse transcribed into cDNA by TCR constant region C-specific primers. The following systems take the number of reactions of a sample as an example.
[0100] 1.1 cDNA 1st synthesis
[0101] 1) Prepare according to the following system (1 sample)
[0102]
[0103] Primer for TCRC region: TTGATGGCTCAAAACACAGCGA (SEQ ID NO: 1)
[0104] 2) 70°C for 10min, place on ice for 1min, add the following system, and incubate at 42°C for 1min.
[0105]
[0106] 3) Add 1 μL Superscript II, react at 42°C for 50 minutes, and react at 70°C for 15 minutes.
[0107] 4) Add 1 μL RNase mix and incubate at 37°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com