CRISPR/Cas9-based novel method for achieving multigene knockout in low-transfection-efficiency cell line
A technology of transfection efficiency and cell lines, applied in the field of molecular biology, can solve problems such as sgRNA and Cas9 off-target effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1: Design, synthesis and vector construction of sgRNA targeting Ecm1 and Pgrn gene exons
[0055] (1) Select the Exon corresponding to the region where the gene mainly functions, and the length is about 400-1200bp;
[0056] (2) Find all NGGs and their first 12 bases in the exon region and perform Blast at NCBI to screen out the sequence that exactly matches the target sequence and the only one (if there is no NGG that meets the requirements, look up the CCN in reverse), reducing the potential off-target sites;
[0057] In this example, sgRNAs for Ecm1 exon 6 and Ecm1 exon 7 and sgRNAs designed for Pgrn exon 5 and Pgrn exon 6 are designed, wherein: the sgRNA sequence for Ecm1 exon 6 is: ggatggcttc ccccctggg (Sequence Listing1); the sgRNA sequence for Ecm1 exon 7 is: agctactgac cccctacaa (Sequence Listing2), and the sgRNA sequence for Pgrn exon 5 is: cgtgctgtgt tatggtcga (Sequence Listing 3); the sgRNA sequence for Pgrn exon 6 is: cggtgccttc tgcgacc (sequence li...
Embodiment 2
[0058] Example 2: Construction and packaging of lentivirus targeting Ecm1 and Pgrn genes
[0059] (1) Cloning two loxP sites in the same direction into the pCDH-CMV-MCS-EF1α-Puro vector to obtain the pCDH-CMV-loxp-MCS-loxp-EF1α-Puro vector;
[0060] (2) Cloning the tandem Ecm1 and Pgrn sgRNA expression components and the Cas9 gene into the pCDH-CMV-loxp-MCS-loxp-EF1α-Puro lentiviral vector to obtain a lentivirus expressing multiple PGRN and ECM1 sgRNA expression components and the Cas9 gene simultaneously Vector pCDH-CMV-loxp-Cas9-Ecm1&Pgrn sgRNAs-loxp-EF1α-Puro (such as figure 1 shown);
[0061] (3) Clone the Cre gene into the pCDH-CMV-MCS-EF1α-Neo vector to obtain the lentiviral vector pCDH-CMV-Cre-EF1α-Neo expressing the Cre protein (such as figure 2 shown);
[0062] (4) The lentiviral vectors pCDH-CMV-loxp-Cas9-Ecm1&PgrnsgRNAs-loxp-EF1α-Puro, pCDH-CMV-Cre-EF1α-Neo were co-transfected with lentiviral packaging plasmids into HEK293T cells for lentiviral packaging, and th...
Embodiment 3
[0063] Example 3: Using lentivirus to specifically knock out Ecm1 and Pgrn genes
[0064] Taking the targeting of the Ecm1 and Pgrn genes of the breast cancer cell line MDA-MB-231 as an example, the process is as follows:
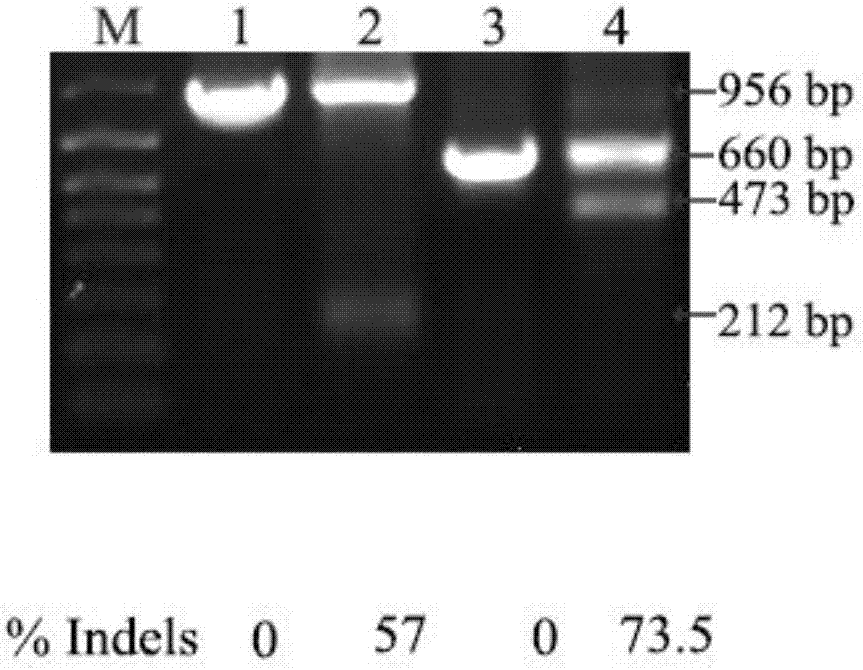
[0065] (1) MDA-MB-231 cells were cultured in 60mm dishes, infected with lentivirus Lenti-Cas9-sgRNAs, added Puromycin (1.0mg / mL) after infection for 3-4 days to obtain MDA integrating Ecm1 and Pgrn sgRNA with Cas9 -MB-231 cells; collect the MDA-MB-231 cell genome integrating Ecm1 and Pgrn sgRNA and Cas9, use primers hEcm1test for sequence aggtaccgaa cgccagctcc atttg (sequence listing 5), hEcm1test back sequence aagatctggccttccatgta caggtgtg (sequence listing 6), hPgrn test for sequence aggtacctgtgtgatgggggagtcacctt (sequence listing 7) and hPgrn test back sequence aagatctgctggctccagcc cctcactca (sequence listing 8) were amplified by PCR and denatured and annealed (such as image 3 , where line 2 is hEcm1, line 4 is hPgrn PCR amplification product, line 1 is...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com