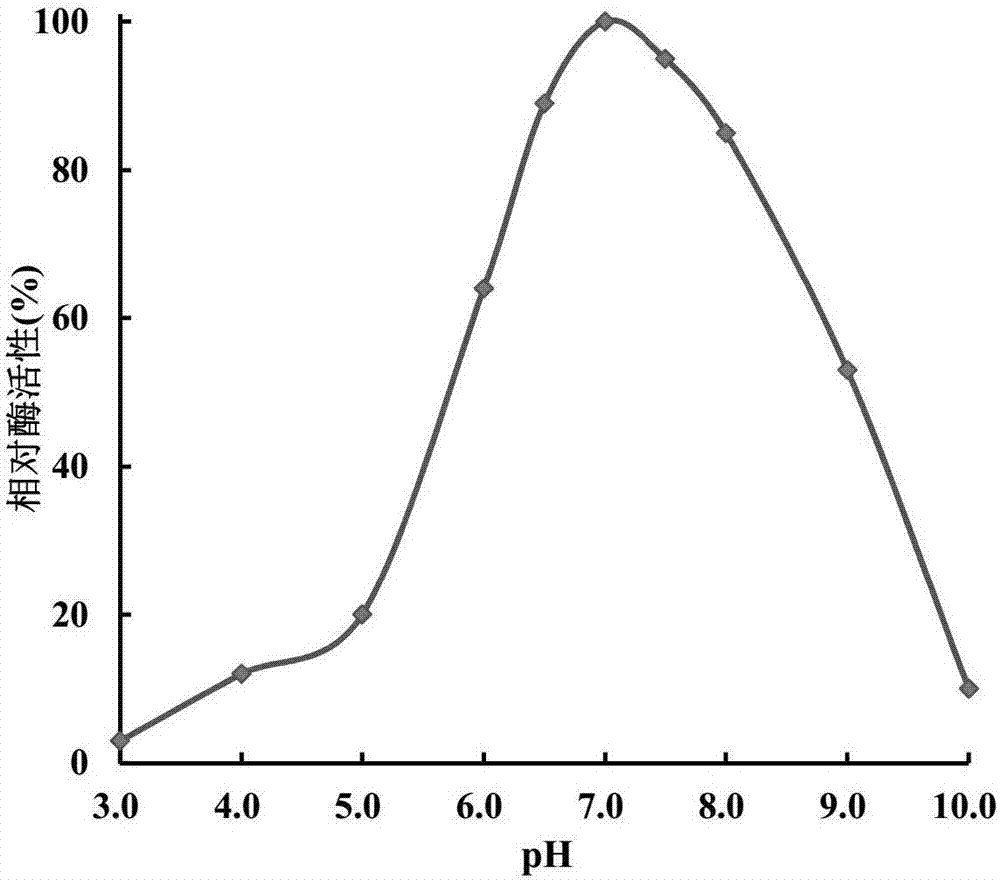
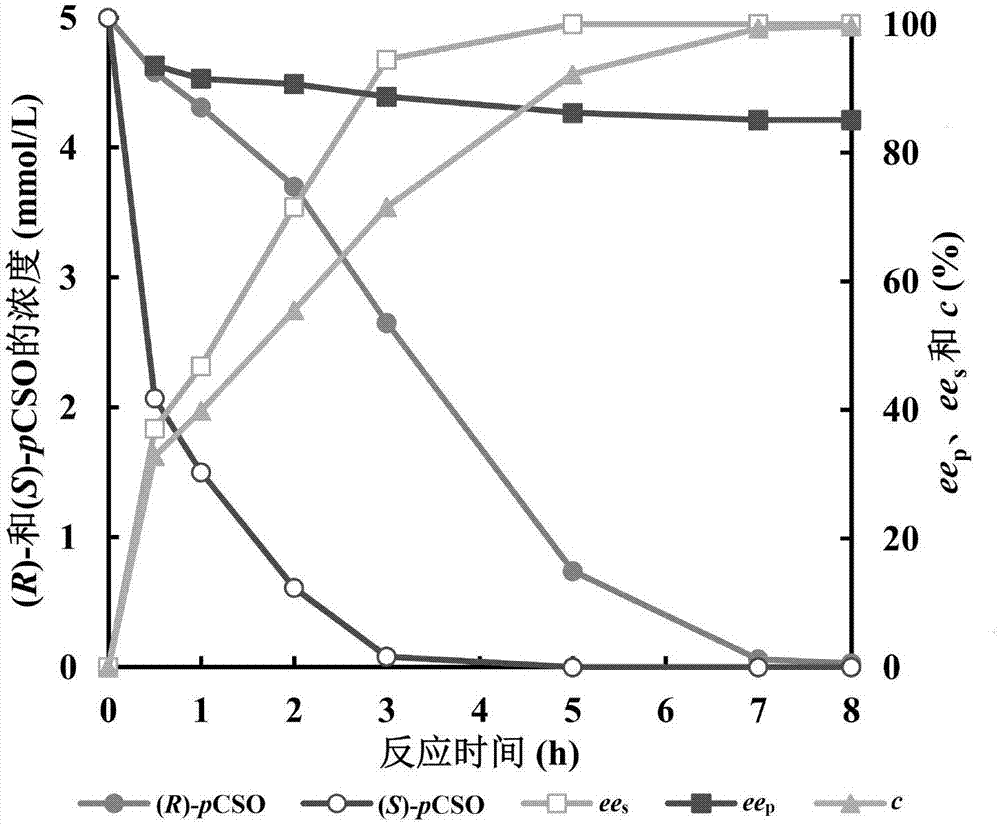
Engineering strain for expressing epoxide hydrolase of kidney beans, and application of engineering strain
A technology of epoxide and hydrolase, applied in the field of bioengineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Obtaining of Phaseolus epoxide hydrolase gene and construction of expression plasmid
[0028] Specific primers were designed according to the cDNA sequence published by NCBI, PvEH2-F: CATATGATGGAGGGAATACACAGCACAAAG, containing Nde I restriction site, PvEH2-R: CTAGAGTCAAAACTTGTTGATAAAATCAT AAAATGT, containing Not I restriction site. Total RNA was extracted from bean germ. The first strand of cDNA was synthesized by reverse transcription using the total RNA of kidney bean as a template and Oligo dT-Adaptor as a primer; using this strand as a template, PvEH2-F and M13Primer M4 as primers, the first round of PCR was performed: denaturation at 94°C for 2 minutes, 30 Cycle (94°C for 30s, 50°C for 30s, and 72°C for 70s); then use the first-round PCR product as a template and PvEH3-F and PvEH3-R as primers to perform a second round of PCR: denaturation at 94°C for 2 min, 30 cycles ( 94°C for 30s, 52°C for 30s and 72°C for 70s), and 72°C for 10min. The PCR product wa...
Embodiment 2
[0029] Induced expression of embodiment 2 recombinant bacteria
[0030] Inoculate a single colony of E.coli BL21-pveh3 in 2 mL of LB medium containing 100 μg / mL kanamycin, and culture overnight at 37°C and 220 r / min; take 2 mL of the culture medium and transfer it to 100 mL of the same medium, Grow to OD 600 When the concentration is 0.6-0.8, add IPTG (final concentration 0.05mmol / L) and induce at 25°C for 8h. Collect the thalline, use 5mL potassium phosphate buffer solution (K 2 HPO 4 -KH2 PO 4 , 100mmol / L, pH 7.0) to obtain a bacterial suspension with a bacterial concentration of 200mg / mL.
Embodiment 3
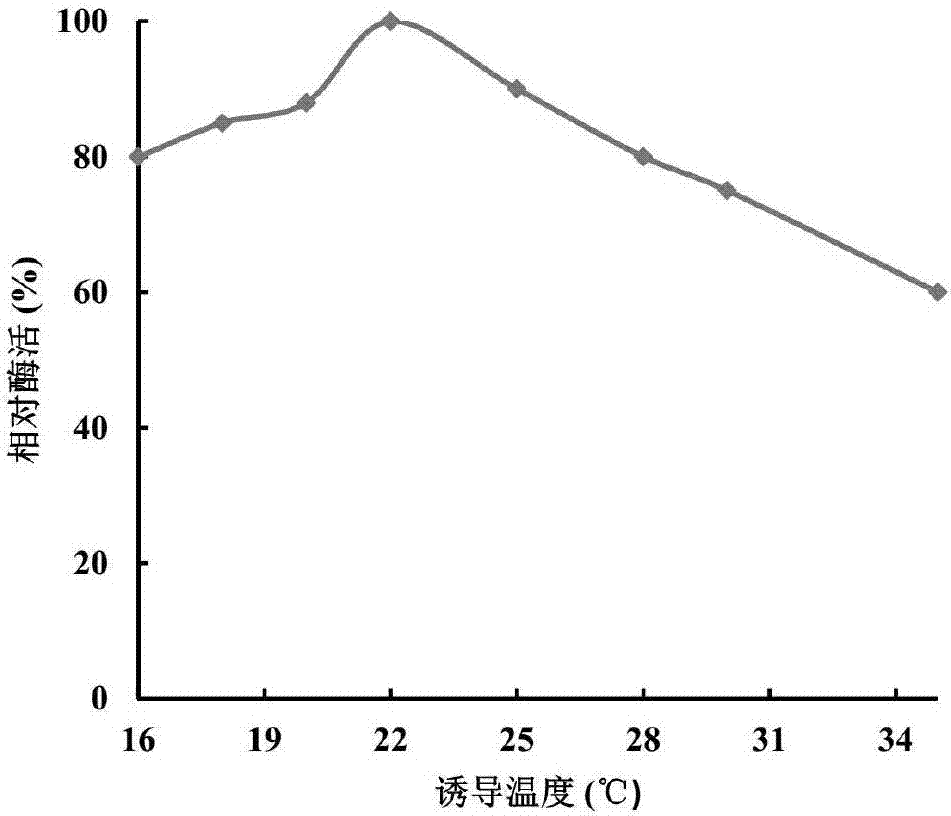
[0031] Induction expression temperature optimization of embodiment 3 recombinant bacteria
[0032] According to the method of Example 2, the difference is that the induction temperature is adjusted to 16-35°C. Such as figure 1 As shown, when the temperature increased from 16°C to 22°C, the relative enzyme activity also increased. When the temperature continued to increase, the relative enzyme activity began to decrease, and when the temperature was 35°C, the relative enzyme activity was only 60% of that at 22°C. It shows that the optimum induction temperature is 22℃.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com