Construction method and application of formate dehydrogenase engineering strain
A technology of formate dehydrogenase and engineering strains, applied in the direction of microorganism-based methods, applications, genetic engineering, etc., can solve problems such as inability to meet market demand, and achieve good industrial applicability and high enzyme activity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Construction of Strain BL21(DE3)-fdh
[0033] Extracting the genome of Candida boidinii as a template, and using a degenerative primer pair as primers for PCR amplification to obtain fdh gene fragments;
[0034] The nucleotide sequence of the upstream degenerate primer is shown in SEQ ID NO.1, and the nucleotide sequence of the downstream degenerate primer is shown in SEQ ID NO.2:
[0035] SEQ ID NO.1: 5'-CCTCTAGAAAGGAGATATAATGAAGATYGTYTTAGTYYTWTATG-3'
[0036] SEQ ID NO.2: 5'-CCGGATCCTTTATTTCTTATCGTGTTTACCG-3'
[0037] The amplification conditions were: pre-denaturation at 94°C for 3 minutes; 30 cycles at 94°C for 30s, 58°C for 30s, and 72°C for 45s; and finally extension at 72°C for 7 minutes.
[0038] The obtained fdh gene fragment was double-digested with endonucleases XbaI and BamHI, and ligated with the plasmid pET28a(+) that was also double-digested with XbaI and BamHI (the ligation product was plasmid pET28a(+)-fdh), and transformed after 8 hours of ligation ...
Embodiment 2
[0042] Induced Expression of Formate Dehydrogenase Activity of Strain BL21(DE3)-fdh and Preparation of Crude Enzyme Solution
[0043] Inoculate BL21(DE3)-fdh into 5 mL of LB liquid medium, add 5 μL of 100 mg / mL kanamycin solution to the medium, and culture in a shaker at 37°C and 180 r / min for 12 to 24 hours . .
[0044] Transfer 3mL of bacterial liquid to 50mL of fresh LB liquid medium, and continue to cultivate at a temperature of 37°C and a shaker speed of 180r / min. After 2-3h, add IPTG solution to a final concentration of 0.5mmol / L for induction , The induction time is 3~5h.
[0045] Centrifuge to collect the bacteria completely, resuspend the bacteria with 20-30mL PBS buffer (0.1M pH7.0), then ultrasonically break (power 200-400W, condition is ultrasonic 3s, intermittent 3s, a total of 10min), centrifuge, put on The clear is the crude enzyme solution.
Embodiment 3
[0047] Determination of Formate Dehydrogenase Activity of Strain BL21(DE3)-fdh
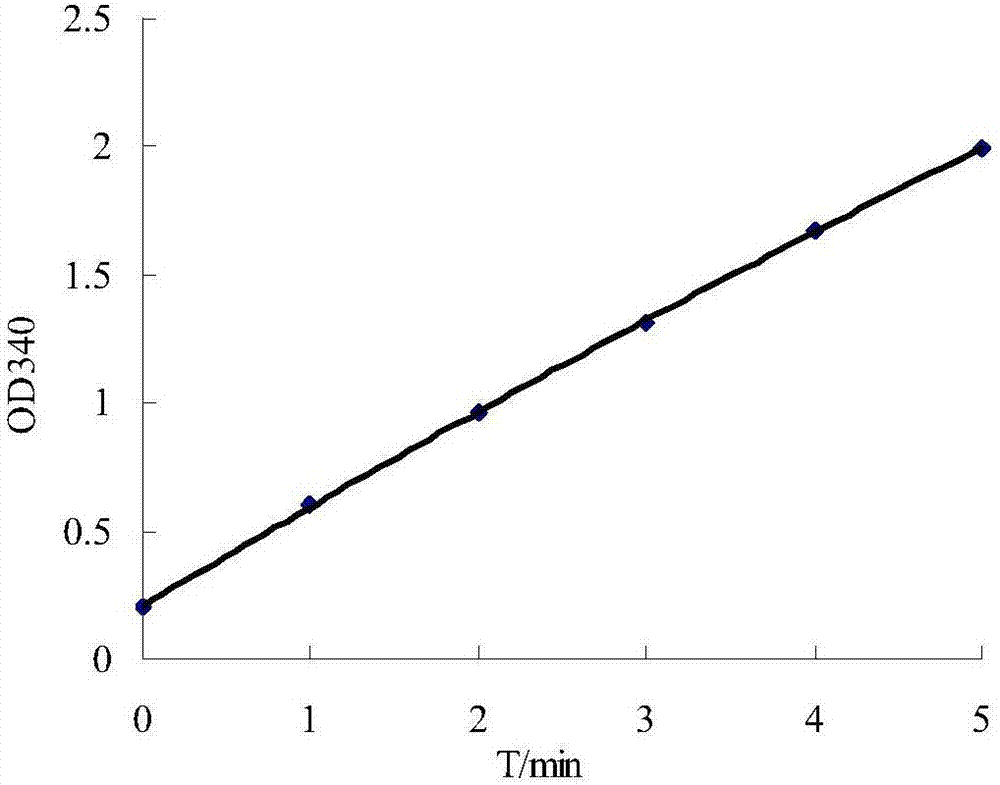
[0048] Enzyme activity assay system: the reaction solution is 0.2mL NAD (16.7mM), 0.2mL sodium formate (1670mM) and 1.4mL PBS buffer (pH7.5, 10mM, containing 4mM DTT), add 0.2mL of diluted crude enzyme solution After mixing, place it in a spectrophotometer immediately, measure the absorbance value A1 at 340nm, react at room temperature (20°C) for 5 minutes, measure the absorbance value A2 at 340nm, A2-A1 is used to calculate the enzyme activity, and the curve of the absorbance value with time is as follows figure 2 shown.
[0049]Enzyme activity is defined as: at pH 7.5 and 30°C, the amount of enzyme required to generate 1 μmol NADH per minute is 1 enzyme activity unit (the molar extinction coefficient of NADH is 6220L·mol -1 cm -1 ).
[0050] The calculated enzyme activity is 0.6331U / mL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com