Method and kit for detecting xanthomonas oryzae pv. oryzae by use of digital-PCR (polymerase chain reaction)
A rice bacterial blight and kit technology, which is applied in the field of plant protection and molecular biology detection, can solve the problems of unreliable diagnosis, time-consuming, false positive and other problems of samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1 is used for the design of the primer pair and specific probe combination of rice bacterial blight digital PCR detection
[0031] According to the rhs family gene of Xanthomonas oryzae, the following 4 primer pairs and specific probe combinations for digital PCR detection of Xanthora oryzae were designed:
[0032] Upstream primer Xoo119F: 5′-AACGGATCGATGGACTTTACG-3′
[0033] Downstream primer Xoo119R: 5′-AATGTGCACTTCCGCTATGA-3′
[0034] Specific probe Xoo119P: 5'-FAM-TTCGTGTGAGGTTACCCTGC-TAMRA-3'
[0035] The 5' end of the probe Xoo119P has a FAM fluorescent label, and the 3' end has a TAMRA fluorescent label.
[0036] Upstream primer Xoo114F: 5′-GATCCGCTCGGTCGCAATGTG-3′
[0037] Downstream primer Xoo114R: 5′-GGCTCGCCCCAGTCCGTCGTA-3′
[0038] Specific probe Xoo114P: 5'-FAM-TGCAGGGTAACCTCACACGAATGG-TAMRA-3'
[0039] The 5' end of the probe Xoo114P has a FAM fluorescent label, and the 3' end has a TAMRA fluorescent label.
[0040] Upstream primer Xoo105F: ...
Embodiment 2
[0049] Example 2 Establishment of digital PCR detection method for bacterial blight of rice
[0050] 1. Preparation of reaction system Reaction system 20ul, consisting of 2×SuperMix 10ul, primer Xoo119F1ul, primer Xoo119R 1ul, probe Xoo119P 0.5ul, genomic DNA of the sample to be tested 2ul and ddH 2 O 5.5ul, genomic DNA in the blank control with ddH 2 O instead.
[0051] 2. After completing step 1, transfer the reaction system to the system sample hole of the digital PCR droplet generation card, and add 70ul of digital PCR reaction oil to the reaction oil sample hole, and transfer the digital PCR droplet generation card Transfer to the DropletGenerator micro-droplet generator, start the instrument, and get the micro-droplet system.
[0052] 3. After completing step 2, transfer the droplet system to a 96-well plate and seal the film;
[0053] 4. After completing step 3, place the 96-well plate in a PCR machine for PCR amplification reaction. The reaction program: pre-denatur...
Embodiment 3
[0056] Embodiment 3 carries out specific detection according to the method established in embodiment 2
[0057] Genomic DNA of seven bacterial strains obtained from the NCPPB collection bank was extracted with a bacterial genomic DNA extraction kit.
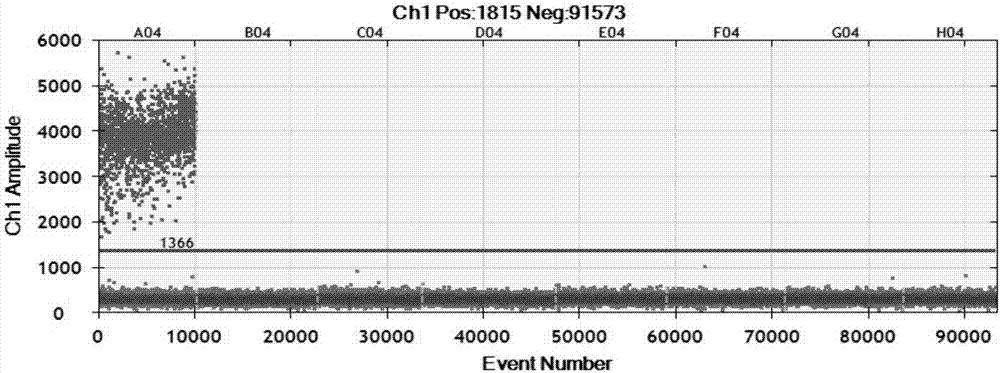
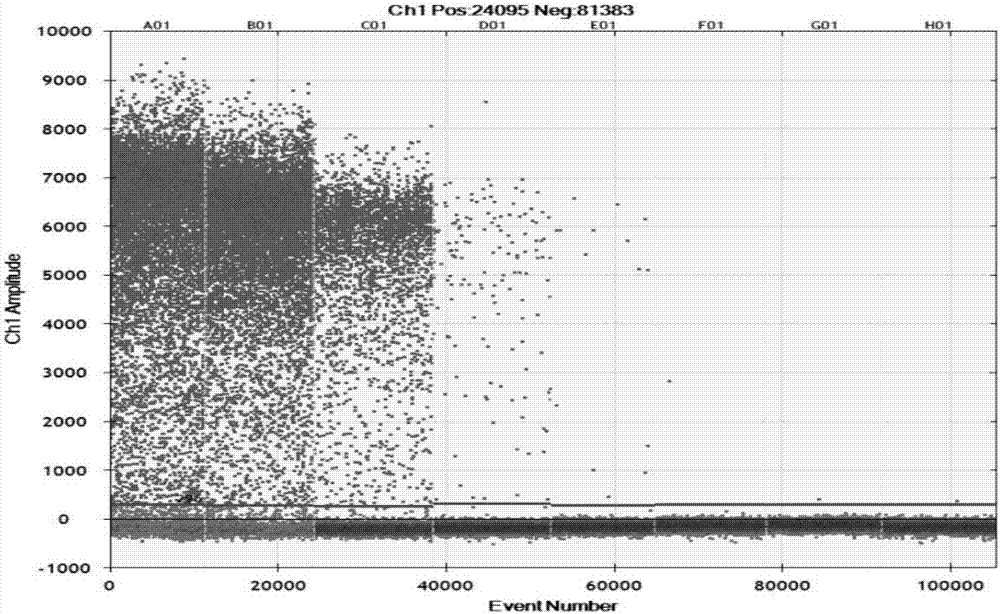
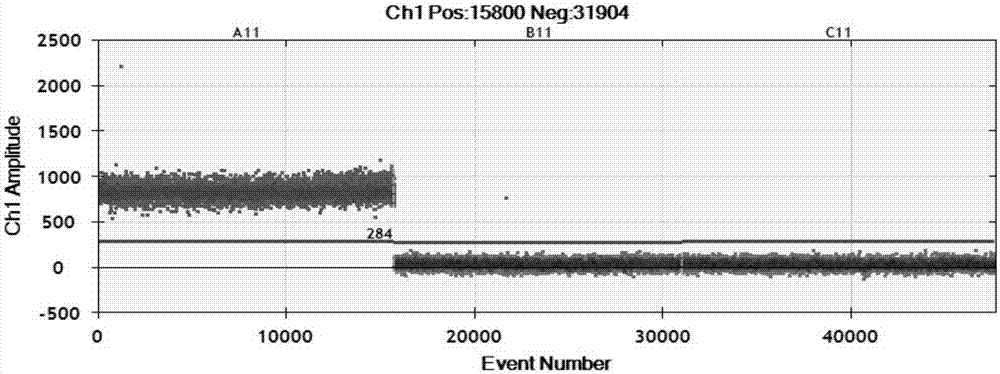
[0058] According to the method of Example 2, specific detection was carried out. See the experimental results figure 1 (A04 is X.oryzae pv.oryzae, B04 is X.oryzae pv.oryzicola, C04 is X.axonopodis pv.citri, D04 is X.axonopodispv.vesicatoria, E04 is B.glumae, F04 is A.avenae subsp.avenae , G04 is P. agglomerans, H04 is blank control). The results showed that only X.oryzae pv.oryzae produced positive micro-droplets, and other bacterial strains and blank controls did not produce positive micro-droplets (at this time, the threshold value as judgment was 1366, and micro-droplet instruments below the threshold value were judged as negative, and those above the threshold value were judged as negative. The microdrop instrument judged ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com