Set of base editing system based on suppurative streptococcus and application of set of basic group editing system in gene editing
A Streptococcus pyogenes, base editing technology, applied in the field of molecular biology, can solve problems such as hindering the application of base editing technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Preparation method of base editing system components based on Streptococcus pyogenes
[0049] Figure 1~4 Expression vector map of a more precise base editing system for four species of Streptococcus pyogenes.
[0050] This example provides a preparation method for base editing system components rAPOBEC1:Cas9:UGI mRNA and gRNA based on Streptococcus pyogenes.
[0051] 1. Preparation of Streptococcus pyogenes rAPOBEC1:Cas9:UGI mRNA, the method is as follows:
[0052] (1) Prepare the transcription vector of HF1-BE2, HF1-BE3, HF2-BE2 or HF2-BE3, the sequence is shown in SEQ ID NO.3, SEQ ID NO.4, SEQ ID NO.5, SEQ ID NO.6 .
[0053] (2) Prepare the transcription template of Streptococcus pyogenes APOBEC1:Cas9:UGI comprising T7 promoter:
[0054]Use KpnI to cut the transcription vector of HF1-BE2, HF1-BE3, HF2-BE2 or HF2-BE3, and then use the PCR product purification kit (Axygen) to perform column purification on the digestion product, and then use nuclease-free...
Embodiment 2
[0066] Example 2 Preparation of base editing system components rAPOBEC1:Cas9:UGI mRNA and gRNA based on Streptococcus pyogenes
[0067] Specifically, the operation procedure of the preparation method of Streptococcus pyogenes APOBEC1:Cas9:UGI mRNA, APOBEC1:Cas9:UGI protein and gRNA described in the above-mentioned embodiment 1 is as follows:
[0068] 1. Preparation of APOBEC1:Cas9:UGI mRNA and gRNA transcription template DNA:
[0069] (1) Preparation of Streptococcus pyogenes APOBEC1:Cas9:UGI mRNA transcription template DNA
[0070] Prepare the transcription vector of HF1-BE2, HF1-BE3, HF2-BE2 or HF2-BE3 by plasmid extraction (independent research and development), and then digest the transcription vector with KpnI, and proceed according to the reaction system shown in Table 1 below:
[0071] Table 1 reaction system
[0072] Element
Dosage
mRNA transcription vector
2000ng
10X NEBuffer 1.1
5μl
Kpn I
5μl
wxya 2 o
Mak...
Embodiment 3
[0133] Example 3 Site-directed mutation of Tyr gene mediated by a more precise base editing system based on Streptococcus pyogenes
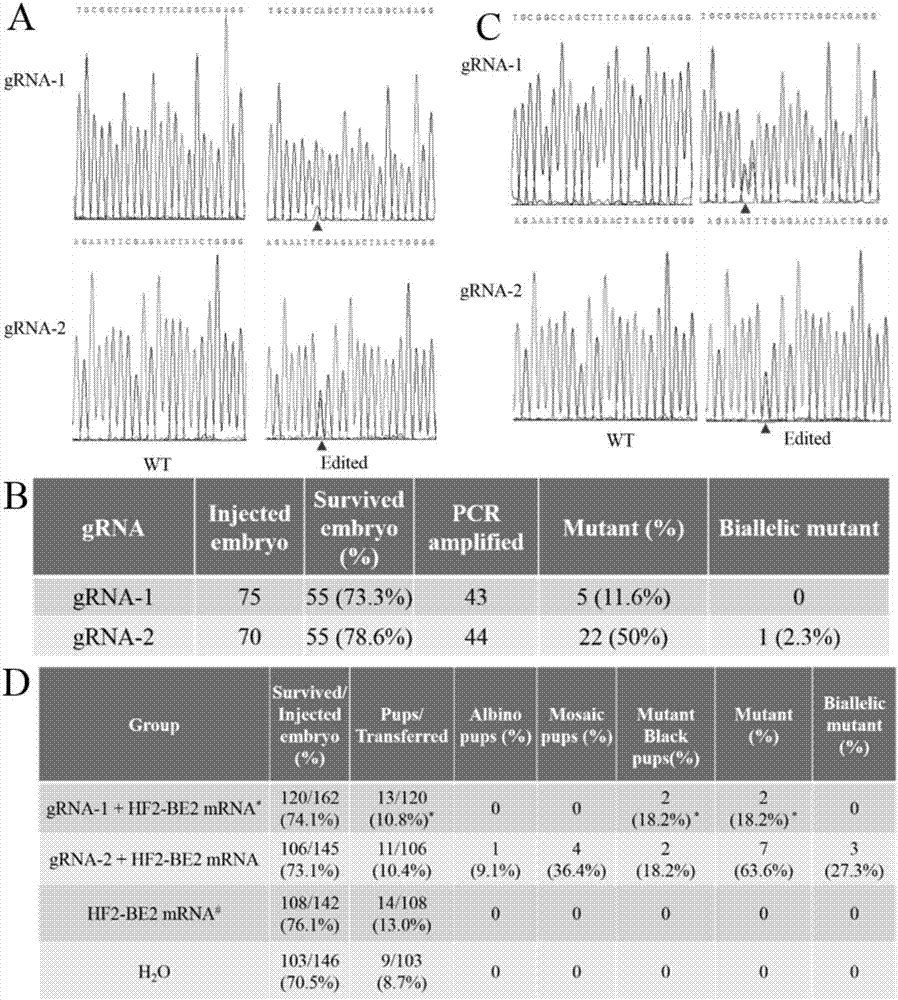
[0134] 1. In order to use a more accurate base editing system based on Streptococcus pyogenes to achieve single-gene site-directed mutation in mouse fertilized eggs, we designed two gRNAs (gRNA-1 and gRNA-2) for the Tyr gene. The site-directed mutation of the Tyr gene will form a stop codon, which will lead to the premature termination of the translation of the Tyr gene, so that the coat color of the pups will change from black to white.
[0135] First, we transcribed Tyr gRNA, then mixed Tyr gRNA (50ng / μl) and rAPOBEC1:Cas9:UGI(HF2-BE2) mRNA (100ng / μl) and injected them into 0.5-day-old mouse fertilized eggs. 48 hours after injection, the efficiency of site-directed mutation was detected. By combining PCR and Sanger sequencing detection, we found that: for gRNA-1, 11.6% of the mouse embryos were mutated, and for gRNA-2, 50% of the embryos were m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com