Multiplex real-time fluorescence quantitative PCR detection kit for transgenic rapeseed RF1, Oxy235 and RF3 and application
A real-time fluorescent quantitative, oxy235-f technology, applied in recombinant DNA technology, DNA/RNA fragments, etc., can solve problems such as low efficiency, achieve high throughput, low cost, and save experimental samples and drugs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Establishment of multiple real-time fluorescent quantitative PCR detection methods for transgenic rapeseed RF1, Oxy235, and RF3
[0042] (1) Synthesize the following primers and probes:
[0043] Rapeseed cruciferinA (CruA) gene in National Standard Announcement No. 2031 of the Ministry of Agriculture-9-2013 "Detection of Rapeseed Internal Standard Gene Qualitative PCR Method for Components of Transgenic Plants and Their Products" was used as the internal standard gene. According to the flanking sequence information of RF1, Oxy235, and RF3 published in the GenBank nucleic acid database, the primers were designed and analyzed using Oligo V7.0 software, and synthesized by Sangon Biopower (Shanghai) Co., Ltd.
[0044] Table 1 Screening results of multiplex real-time fluorescent quantitative PCR primers
[0045]
[0046]
[0047] RF1-F: 5'-AATTTGGCCTGTAGACCT-3', its nucleotide sequence is shown in SEQ ID NO.1;
[0048] RF1-R: 5'-TACACGCGACTCATCATCC-3', its ...
Embodiment 2
[0070] Embodiment 2 specificity test
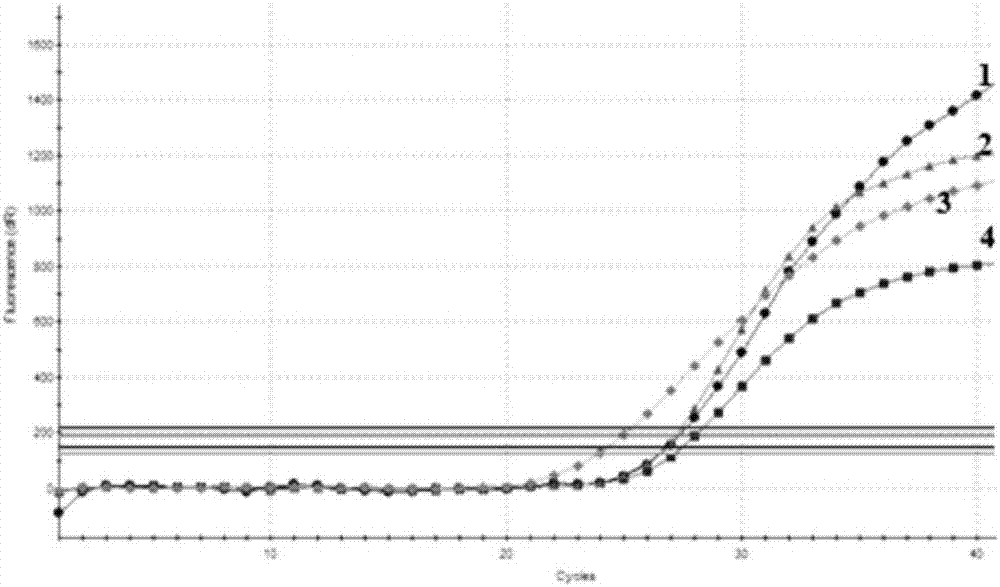
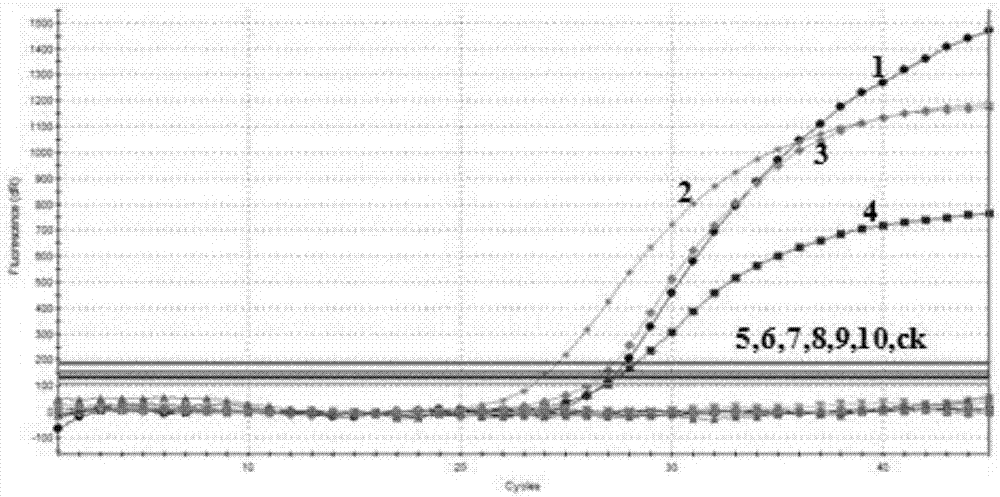
[0071] In order to evaluate and verify the specificity of multiple real-time fluorescent quantitative PCR primers and probes, genomic DNA was extracted from transgenic rapeseed, transgenic maize, transgenic soybean, transgenic rice, transgenic cotton and other transgenic crops with different transformation vectors for verification. The results showed that the expected amplification curve was obtained only in the transgenic rapeseed containing the objective transformation vector.
[0072] Conclusion: by figure 2 The results show that according to the established multiple real-time fluorescent PCR method for specificity testing, CruA, Oxy235, RF3 and RF1 have expected amplification curves within 36 Ct values, indicating that this method can detect CruA, Oxy235, RF3 and RF1 target components.
Embodiment 3
[0073] Embodiment 3 sensitivity test
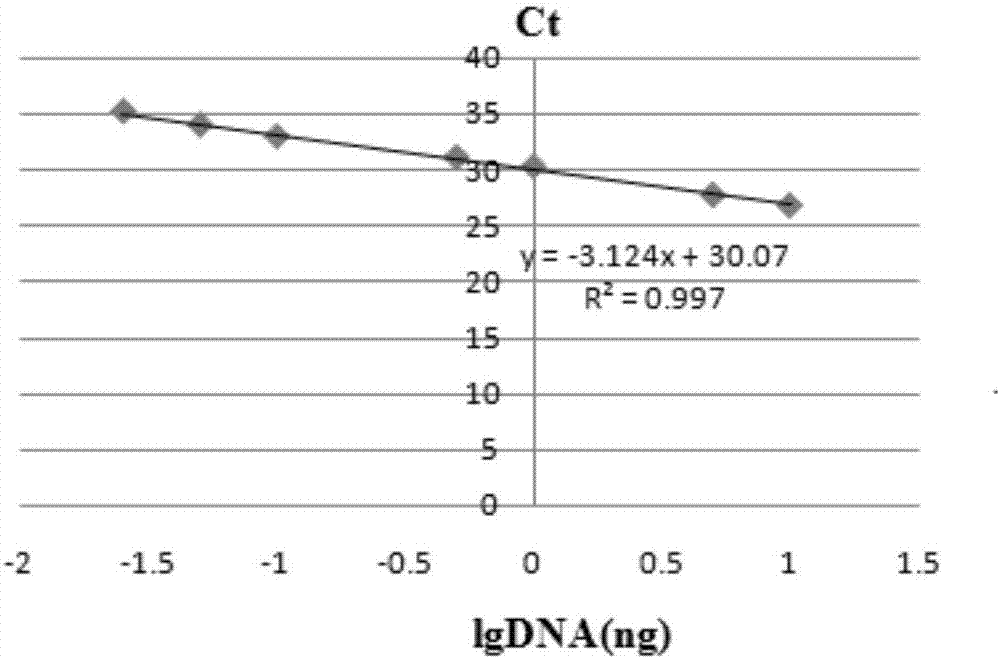
[0074] Genomic DNA standard substances of transgenic rapeseed with three different transformation vectors were diluted with non-transgenic rapeseed genomic DNA, mixed thoroughly, and prepared into mass fraction ratios of 10%, 5%, 1%, 0.5%, 0.1%, 0.05%, and 0.025% and 0.01% etc. a series of genomic DNA templates. 100 ng each was taken as a template (20 μL system) for real-time fluorescent PCR test, and each diluted sample was set for 3 replicates. Find the Ct value of 3 repetitions SD and RSD. The standard curve and regression equation between Ct value and template amount were fitted to determine the linearity and sensitivity of the established multiple real-time fluorescent PCR method.
[0075] According to the provisions of the European Network of Transgenic Laboratories on the detection method of genetically modified components (2011), it is required that the slope of the standard curve between the Ct value of real-time fluorescent ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com