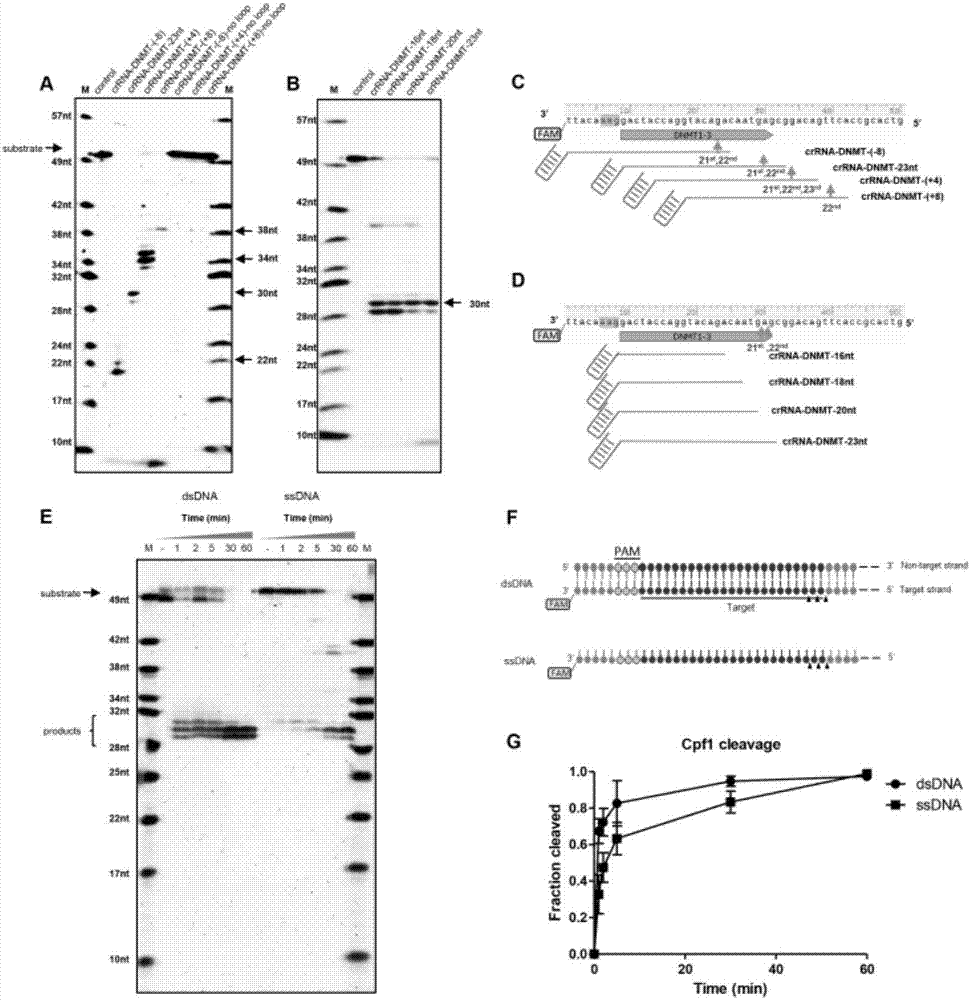
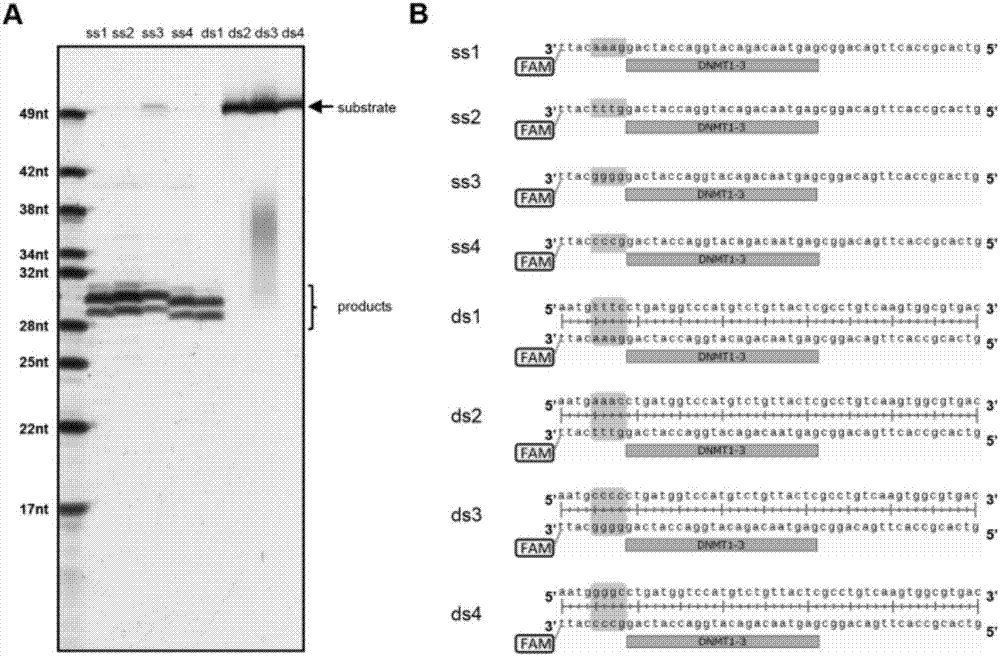
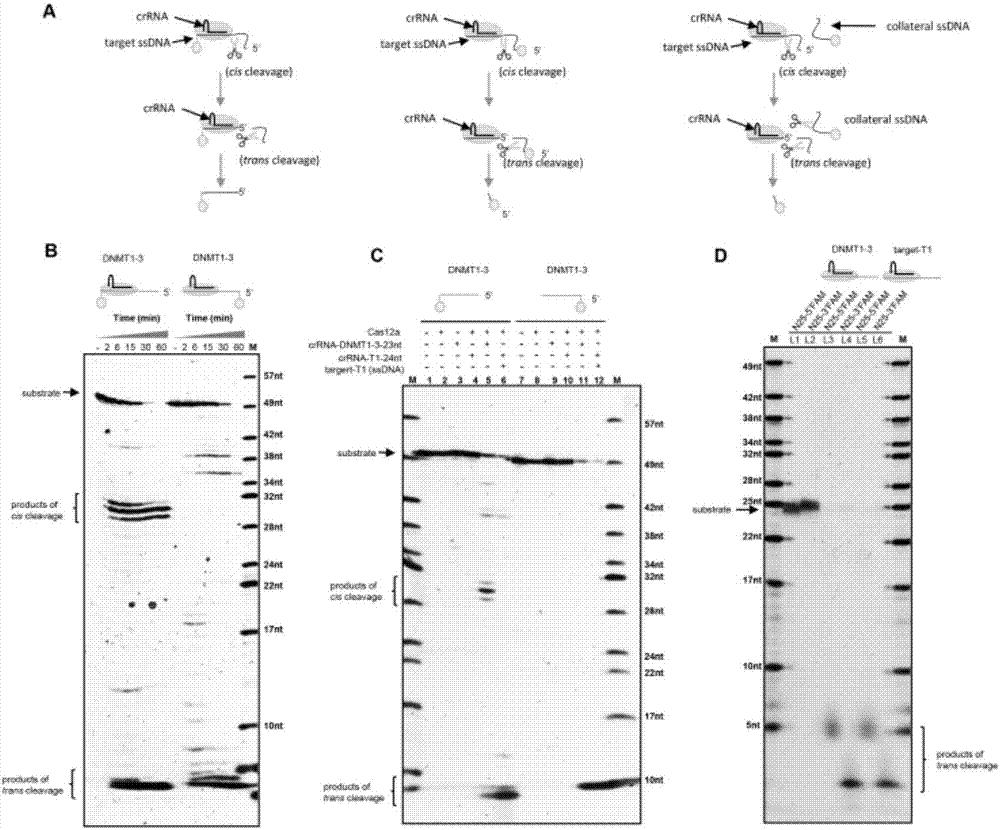
An application of a Cas protein, and a method and kit for detecting a target nucleic acid molecule
A detection method and target nucleic acid technology, applied in biochemical equipment and methods, microbial measurement/inspection, genetic engineering, etc., can solve problems such as increasing operational difficulty and achieve high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 Cas12a protein detects the target of single-stranded DNA (ssDNA) (probe FAM label)
[0077] Single-stranded DNA (target-T1-R) was selected as the target sequence to test the response value of different Cas12a proteins to its detection.
[0078]1. Preparation of crRNA: First, the transcription template was prepared by annealing T7-crRNA-F and synthetic oligonucleotide T7-T1-24-R, as shown in Table 5. Specifically, paired oligonucleotides (4 μM) were annealed in 1× PCR buffer (Transgen Biotech) in a total volume of 50 μL, followed by an annealing procedure: initial denaturation at 95 °C for 5 min, followed by cooling from 95 °C to 20 °C, using a thermal cycler to decrease 1 °C per minute. crRNA was synthesized using the T7 High Yield Transcription Kit, and the reaction was performed overnight (approximately 16 h) at 37°C. RNA was then purified using the RNA Purification and Concentration Kit, quantified with NanoDrop 2000C (Thermo Fisher Scientific), diluted t...
Embodiment 2
[0081] Example 2 Cas12a protein detects the target of single-stranded DNA (ssDNA) (the probe has HEX, BHQ1 double labeling)
[0082] Single-stranded DNA (target-T1-R) was selected as the target sequence to test the response value of different Cas12a proteins to its detection.
[0083] 1. Preparation of crRNA: First, the transcription template was prepared by annealing T7-crRNA-F with the synthetic oligonucleotide T7-T1-24-R (Table 5). Specifically, paired oligonucleotides (4 μM) were annealed in 1×PCR buffer (TransgenBiotech) in a total volume of 50 μL, followed by an annealing procedure: initial denaturation at 95°C for 5 min, followed by cooling from 95°C to 20°C °C, decreasing 1 °C per minute using a thermal cycler. crRNA was synthesized using the T7 High Yield Transcription Kit, and the reaction was performed overnight (about 16h) at 37°C. The RNA was then purified using the RNA Purification and Concentration Kit, quantified with NanoDrop2000C, diluted to a concentration...
Embodiment 3
[0086] Example 3 Cas12a protein detects the target of double-stranded DNA (dsDNA)
[0087] Double-stranded DNA (target-T1) was selected as the target sequence to test the response value of different Cas12a proteins to its detection.
[0088] 1. Preparation of crRNA: First, the transcription template was prepared by annealing T7-crRNA-F with the synthetic oligonucleotide T7-T1-24-R (Table 5). Specifically, paired oligonucleotides (4 μM) were annealed in 1×PCR buffer (TransgenBiotech) in a total volume of 50 μL, followed by an annealing procedure: initial denaturation at 95°C for 5 min, followed by cooling from 95°C to 20°C °C, decreasing 1 °C per minute using a thermal cycler. crRNA was synthesized using the T7 High Yield Transcription Kit, and the reaction was performed overnight (about 16h) at 37°C. The RNA was then purified using the RNA Purification and Concentration Kit, quantified with NanoDrop2000C, diluted to a concentration of 10 μM and stored in a -80°C refrigerator...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com