Chrysanthemum strigolactone synthetase gene CmCCD8 and application thereof in changing chrysanthemum flowering phase
A gene, chrysanthemum technology, applied in the field of plant genetic engineering and transgenic breeding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
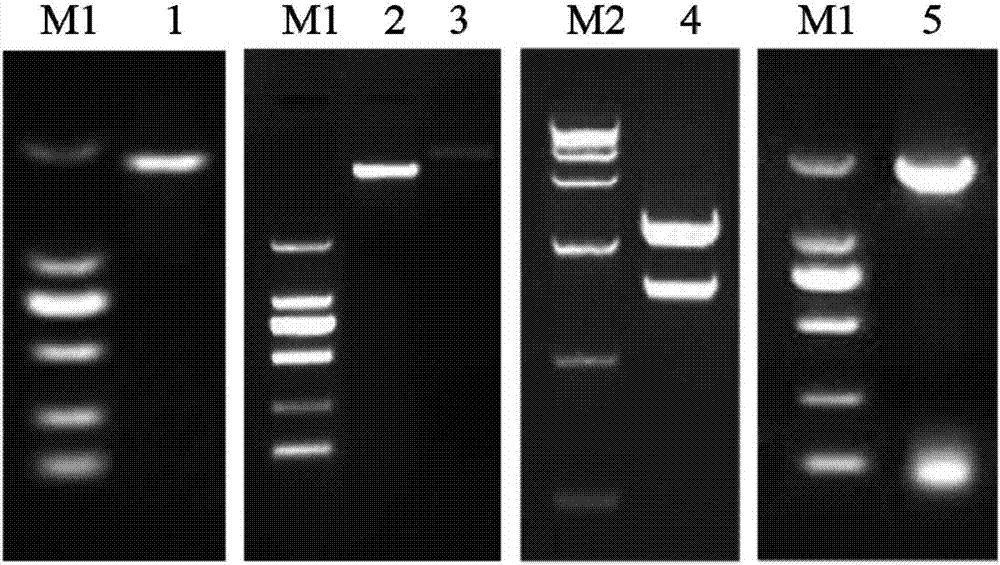
[0085] Example 1. Cloning of CmCCD8 gene
[0086] Take cut chrysanthemum "Shenma" as material, take 0.15g of root system, extract the total RNA of root system according to the operation method of Trizol RNA extraction kit (TaKaRa), and perform reverse transcription according to M-MLV reverse transcription kit (TaKaRa) Obtain cDNA, and use primer 5 software to design specific primers to amplify CmCCD8 according to the sequence information of the gene in the chrysanthemum library;
[0087] Upstream primer CmCCD8-F: 5'-ATGGCTTCCTCCCTACTTTC-3' (SEQ ID NO. 2),
[0088] Downstream primer CmCCD8-R: 5'-TCAACATTTTGGAACCCAACAT-3' (SEQ ID NO.3);
[0089] Use root cDNA as template to carry out PCR reaction, 50μL reaction system: 10×PCR Buffer 5.0μL, CmCCD8-F, CmCCD8-R primers 1.0μL each (20μmol·L -1 ), dNTP mix 4.0μL(2.5mmol·L -1 ), Taq DNAPolymerase 0.2μL, cDNA template 1μL, ddH 2 O 37.8μL; reaction procedure: 95°C pre-denaturation for 5min, then 94°C melting for 45sec, 55°C annealing for 45sec,...
Embodiment 2
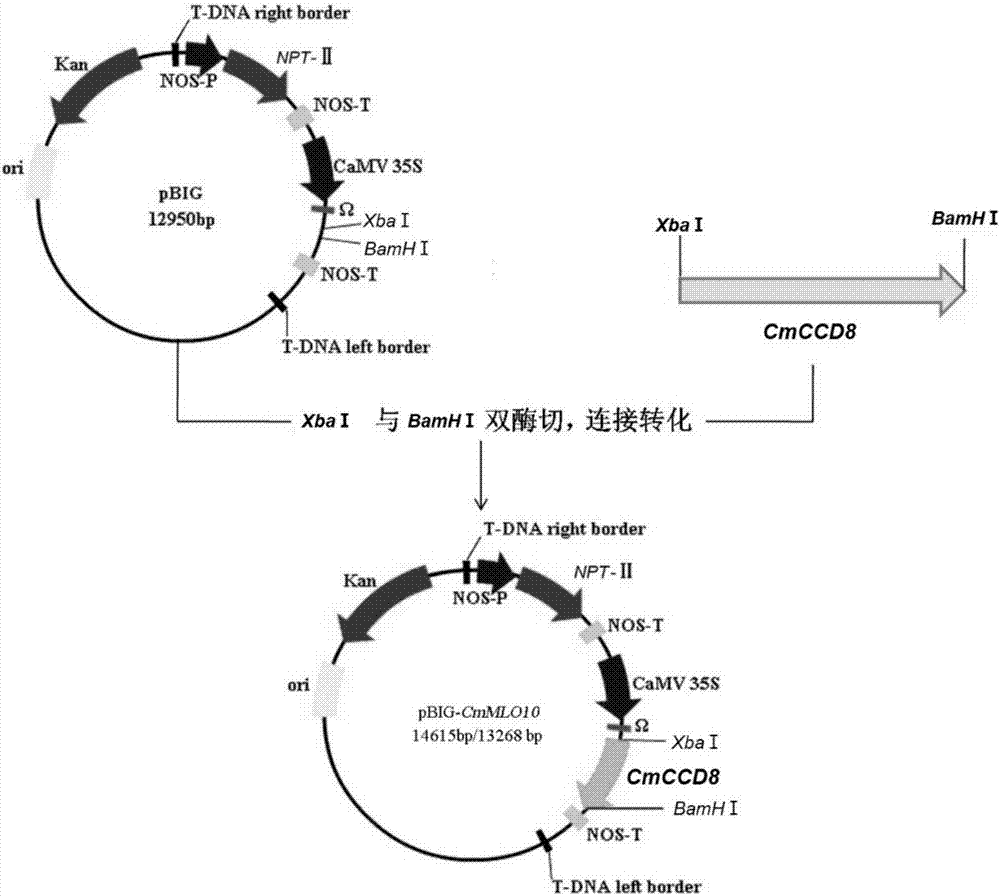
[0090] Example 2. Construction of plant expression vector pBIG-CmCCD8
[0091] Design primers based on the full-length gene sequence of CmCCD8 to perform PCR reaction, introduce restriction sites Xba I and BamH I upstream and downstream of the target gene CmCCD8, respectively, connect the PCR product to the pMD19-T vector, transform DH5α competent cells, and extract the positive plasmid The CmCCD8 fragment obtained by double digestion with Xba I and BamH I of the extracted positive plasmid was ligated with pBIG double digested with Xba I and BamH I, transformed, and the positive plasmid was extracted. Upstream primer CmCCD8-Xba I-F: 5′-GCTCTAGAGCATGGCTTCCTCCCTACTTTC-3′ (SEQ ID NO. 4), downstream primer CmCCD8-BamH I-R: 5′-CGGGATCCCGTCAACATTTTGGAACCCAACAT-3′ (SEQ ID NO. 5);
[0092] Using cut chrysanthemum'shenma' root cDNA as template, using high-fidelity enzyme (PrimeSTAR TM HS DNAPolymerase, TaKaRa) for PCR reaction, 50μL reaction system: 10×HS PCR Buffer 5.0μL, CmCCD8-XbaI-F,...
Embodiment 3
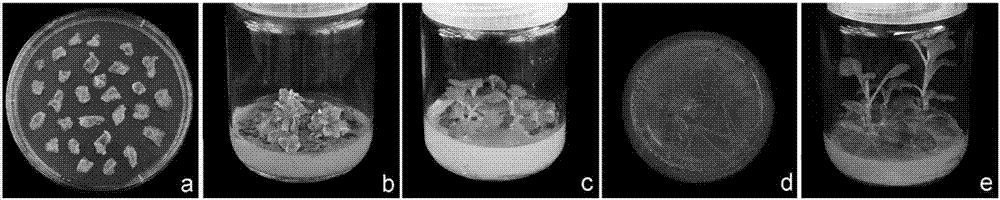
[0094] Example 3. Agrobacterium EHA105 mediates leaf disc method to transform chrysanthemum
[0095] Pick a single colony of EHA105 from YEB (50μg / mL rifampicin) plate, inoculate it in 50mL YEB liquid medium containing 50μg / mL rifampicin, culture at 200rpm and 28℃ to OD value 0.5, then ice bath the bacteria liquid Collect the bacteria by centrifugation for 30min, and suspend in 2mL pre-cooled 100mM CaCl 2 (20% glycerol) solution, 200μL / tube aliquot, set aside.
[0096] Take 10μL pBIG-CmCCD8 vector plasmid, add 200μL competent cells, ice bath for 30min, freeze in liquid nitrogen for 5min, 37℃5min, add 800μL YEB liquid medium, pre-incubate at 28℃200rpm for 4h, and coat the bacterial solution on YEB (50μg / mL rifampicin+50μg / mL kanamycin) solid medium, culture in the dark at 28°C for 2 days, pick a single clone for detection, select positive clones to OD = 0.5, centrifuge, discard the supernatant, and use MS (pH 5.8) The culture solution suspends the precipitate in an equal volume fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com