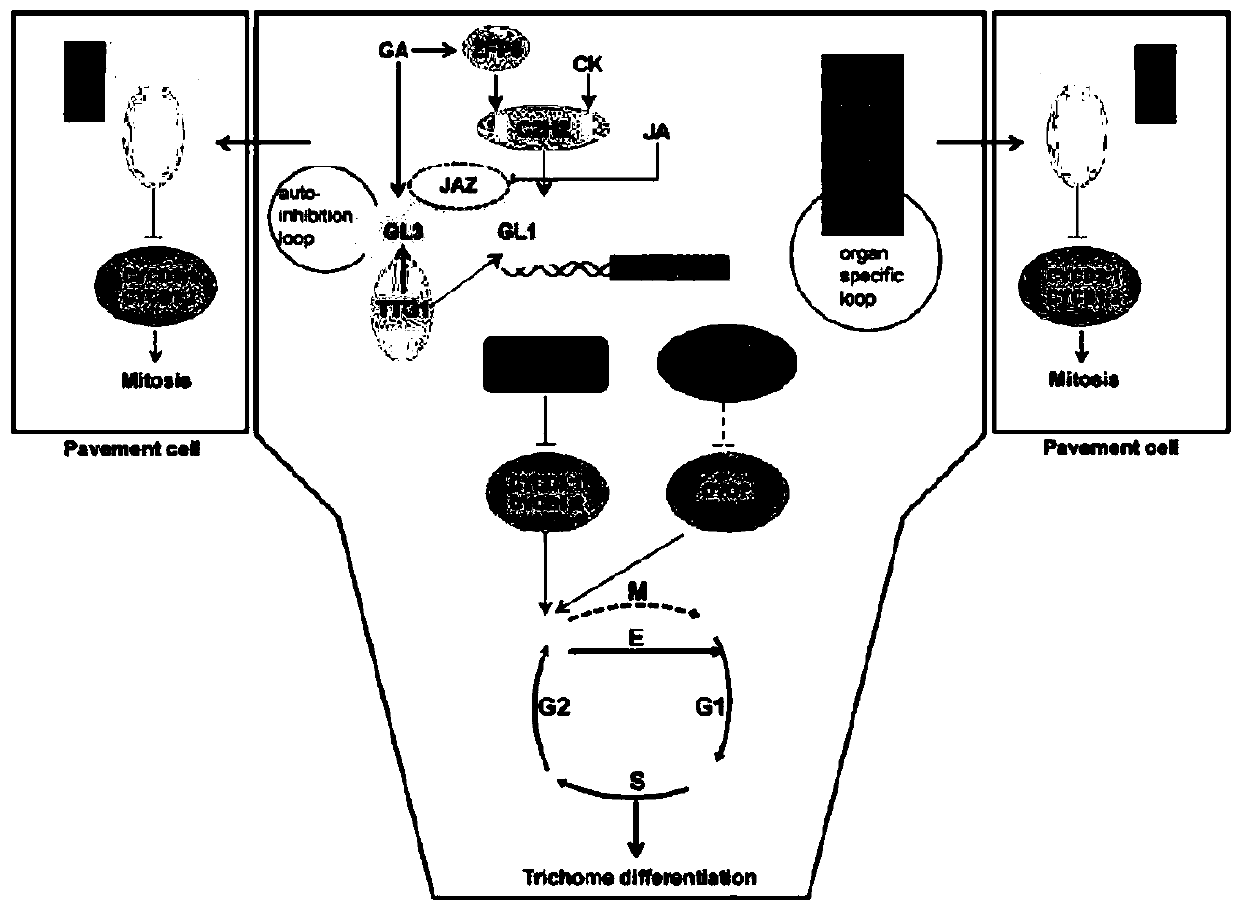
A h gene and h protein regulating the formation of tomato i-type glandular trichomes and their application
A gene and protein technology, applied in the field of H protein, which regulates the formation of tomato type I glandular hairs, can solve the problems such as the lack of thorough research on the regulation mechanism of the regulation gene, and achieve the goal of enhancing the ability of anti-insect and anti-virus, and increasing the density of glandular hairs. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Precise positioning and cloning of the H gene of the present invention
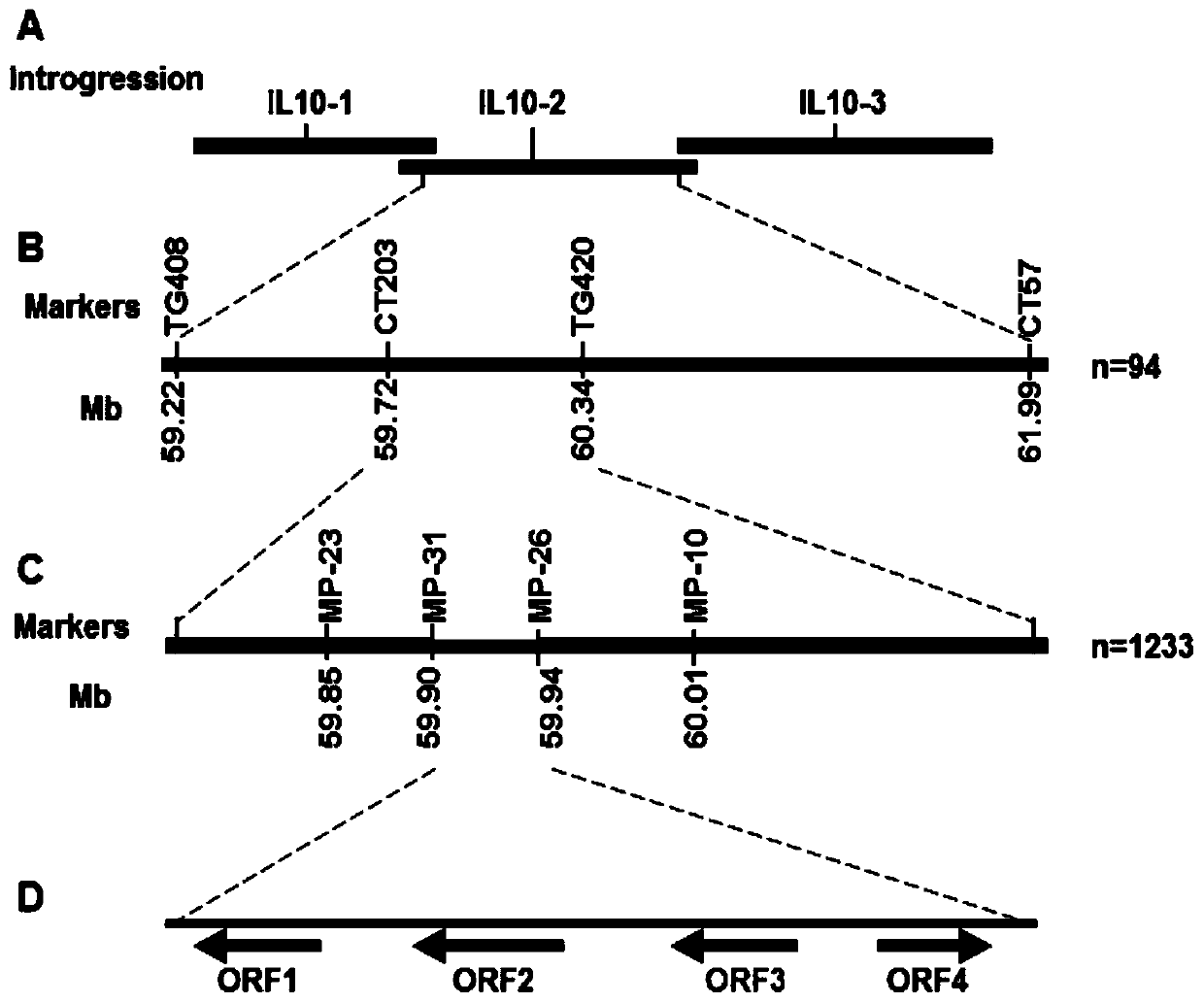
[0040] The phenotype of the h mutant LA3172 showed loss of epidermal hairs ( figure 2 , A is the glandular trichome phenotype of common tomato containing H gene; B is the glandular trichome phenotype of h mutant). In order to clone this key gene, this example uses Ailsa Craig (AC) and IL10-2 (an introgression line constructed by crossing the wild tomato species Panali and cultivar M82) to construct the F2 isolate group map, and screen out the F2 isolate group The genotypes of 94 epidermis-deficient individuals were detected with molecular markers TG408, CT203, TG420 and CT42, and their phenotypes and genotypes were converted into analyzable data for analysis. Linkage analysis showed that the H gene was located between molecular markers CT203 and TG420, with 1 crossover and 3 crossovers, respectively. According to the physical positions of these two markers, the H gene is located on chromosome 10...
Embodiment 2
[0045] Cloning and sequence analysis of H gene
[0046] According to the information of the candidate H gene, PCR-specific primers capable of amplifying the full length of the gene were designed. The forward primer was 5' gcctcgagaacacactcacacacacacaaaatc3' (SEQ ID NO: 19), and the reverse primer was 5' gcggtaccgaaaatagagcagacagaaaaacaa 3' (SEQ ID NO:20). Genomic RNA was extracted from tomato leaves, the RNA was reverse-transcribed into cDNA using a reverse transcriptase kit (purchased from Transgen), and the full-length H gene product was obtained by PCR amplification, which was detected by 1% agarose gel and recovered with a recovery reagent The target fragment was recovered from the kit (purchased from Axgen Company), cloned with the pEasy-Blunt vector (purchased from Transgen Company), and 5 μL of the ligated product was taken for heat shock treatment. Refer to the kit instructions for specific methods, transform Escherichia coli DH5α, and smear it on On the LB solid plat...
Embodiment 3
[0050] Functional verification of the H gene
[0051] Utilize the specific primers (SEQ ID NO:19 and SEQ ID NO:20) of the designed H gene, use high-fidelity DNA polymerase enzyme (purchased from Transgen Company), amplify the full-length cDNA sequence of H gene from Ailsa Craig, After the amplified product was gel-cut and recovered (the recovery kit was purchased from Axgen), it was connected to the pEASY-Blunt intermediate vector (purchased from Transgen), and the correct positive clone was selected by sequencing, and the target clone was digested with XbaI and KpnI. To clone the full-length fragment of gene H into the pEASY-Blunt vector, cut the pMV2 expression vector with XbaI and KpnI at the same time, recover and ligate to obtain the recombinant overexpression vector pMV2-H containing the H gene, see Figure 4 , the vector is driven by the 35S promoter, and the selectable marker gene is Spectinomycin (Spec). The overexpression vector pMV2-H of the H gene was transformed ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com