Application for suppressing expression of gene rsaL in pseudomonas aeruginosa and pseudomonas aeruginosa with suppressed expression of gene rsaL
A Pseudomonas aeruginosa and gene technology, applied in the fields of application, genetic engineering, plant gene improvement, etc., can solve the problems of unstable production performance of strains, easy recovery of physical and chemical mutagenesis, etc., to enhance exercise ability and solve recovery mutations , high yield effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0021] According to a preferred embodiment of the present invention, the rhamnolipid is total rhamnolipid synthesized by Pseudomonas aeruginosa.
[0022] According to another preferred embodiment of the present invention, the rhamnolipid is double rhamnolipid Rha-Rha-C synthesized by Pseudomonas aeruginosa 10 -C 10 and the monorhamnolipid for Rha-C 10 -C 10 .
[0023] In the present invention, the Pseudomonas aeruginosa can be existing Pseudomonas aeruginosa that can synthesize rhamnolipids, but the present invention is particularly suitable for Pseudomonas aeruginosa PAO1. Therefore, according to a preferred embodiment of the present invention, the Pseudomonas aeruginosa is Pseudomonas aeruginosa PAO1. Wherein, the Pseudomonas aeruginosa PAO1 can be obtained through conventional commercial channels, or obtained through other available channels. The Pseudomonas aeruginosa PAO1 is described in detail in the literature Geneticrecombination in Pseudomonas aeruginosa. J Gen M...
Embodiment 1
[0039] This example is used to illustrate the knockout of the rsaL gene
[0040]The gene deleted in the RsaL deletion mutant strain ΔrsaL is located at PA1431 on the Pseudomonas aeruginosa PAO1 genome (www.pseudomonas.com). The construction reference of ΔrsaL is Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nature Protocol, 2015 , 10(11):1820-41. The knockout plasmid is pEX18AP, and the reference is A broad-host-range Flp-FRTrecombination system for site-specific excision of chromosomely-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosamutants.Gene,1998,212(1):77-86 . Primers were designed according to the rsaL gene promoter to amplify its upstream and downstream homologous fragments. The forward primer of the upstream homologous fragment is shown in SEQ ID No: 4 (5'-gccagtgccaagcttgagcattacgaccgggc); the reverse primer is shown in SEQ ID No: 5 (3'-gatcagagcaggcaagatcagagagtaataag); the forward prime...
Embodiment 2
[0043] This example is used to illustrate the comparison of the engineering strain PAO1ΔrsaL provided by the present invention with the starting strain PAO1 swarming exercise ability.
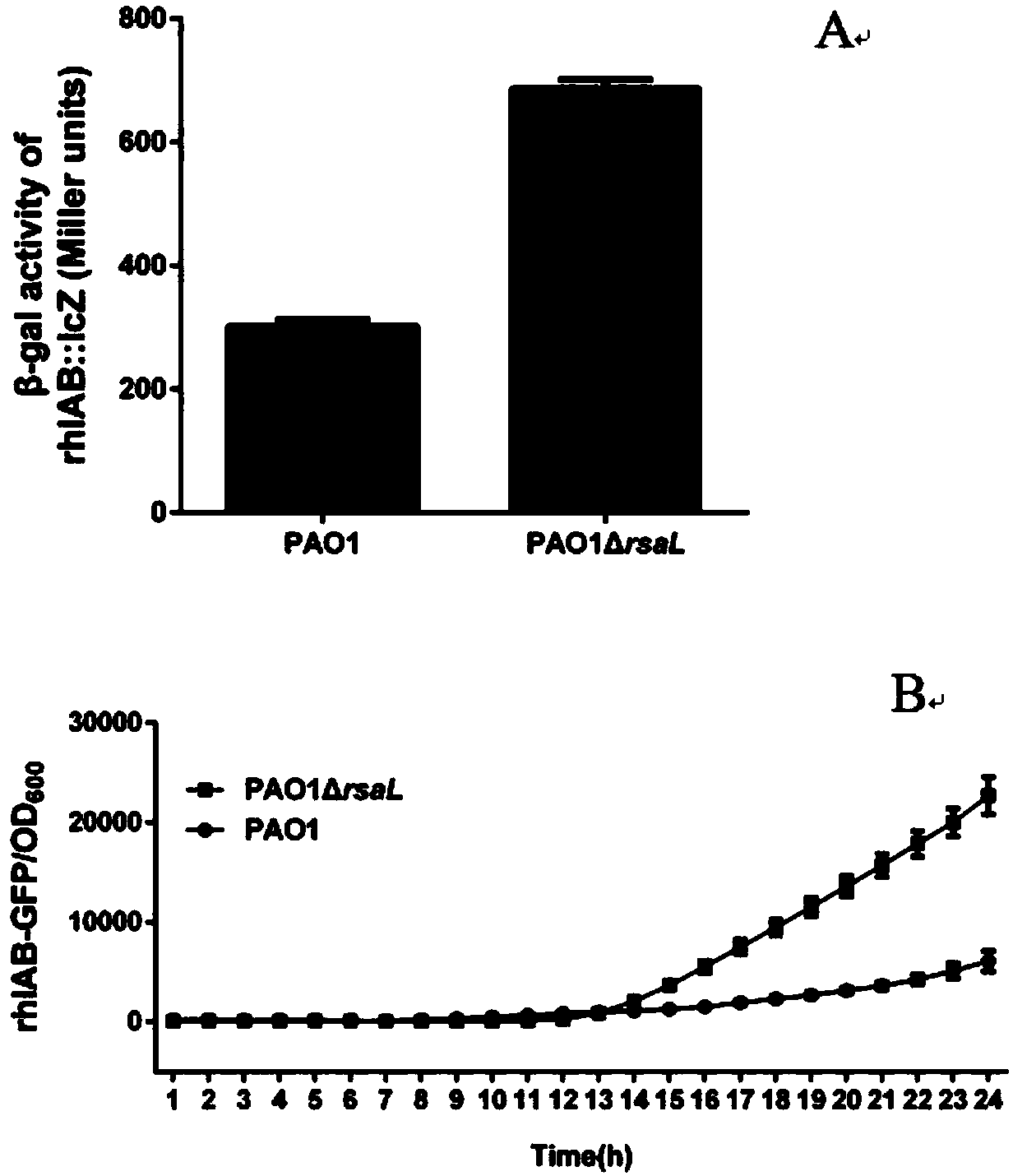
[0044] The activated strains were inoculated into LBNS liquid medium, and cultured with shaking at 37°C overnight. Take 1 μl of the bacterial solution and inoculate it on the surface of the swarming plate (Nutrientbroth 8g / L, D-glucose 5g / L, BactoAgar 5g / L, sterilized at 115°C for 15min), and culture overnight at 37°C. Measure the diameter of the bacterial movement area on each plate, the results are shown in figure 2 .
[0045] From figure 2 It can be seen that the swarming ability of PAO1ΔrsaL (6.9±0.3cm) is significantly greater than that of PAO1 (3.1±0.2cm).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com