Construction method and application of single-cell methylation sequencing library
A library construction, single-cell technology, applied in chemical libraries, biochemical equipment and methods, combinatorial chemistry, etc., can solve problems such as affecting library coverage and inability to perform sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
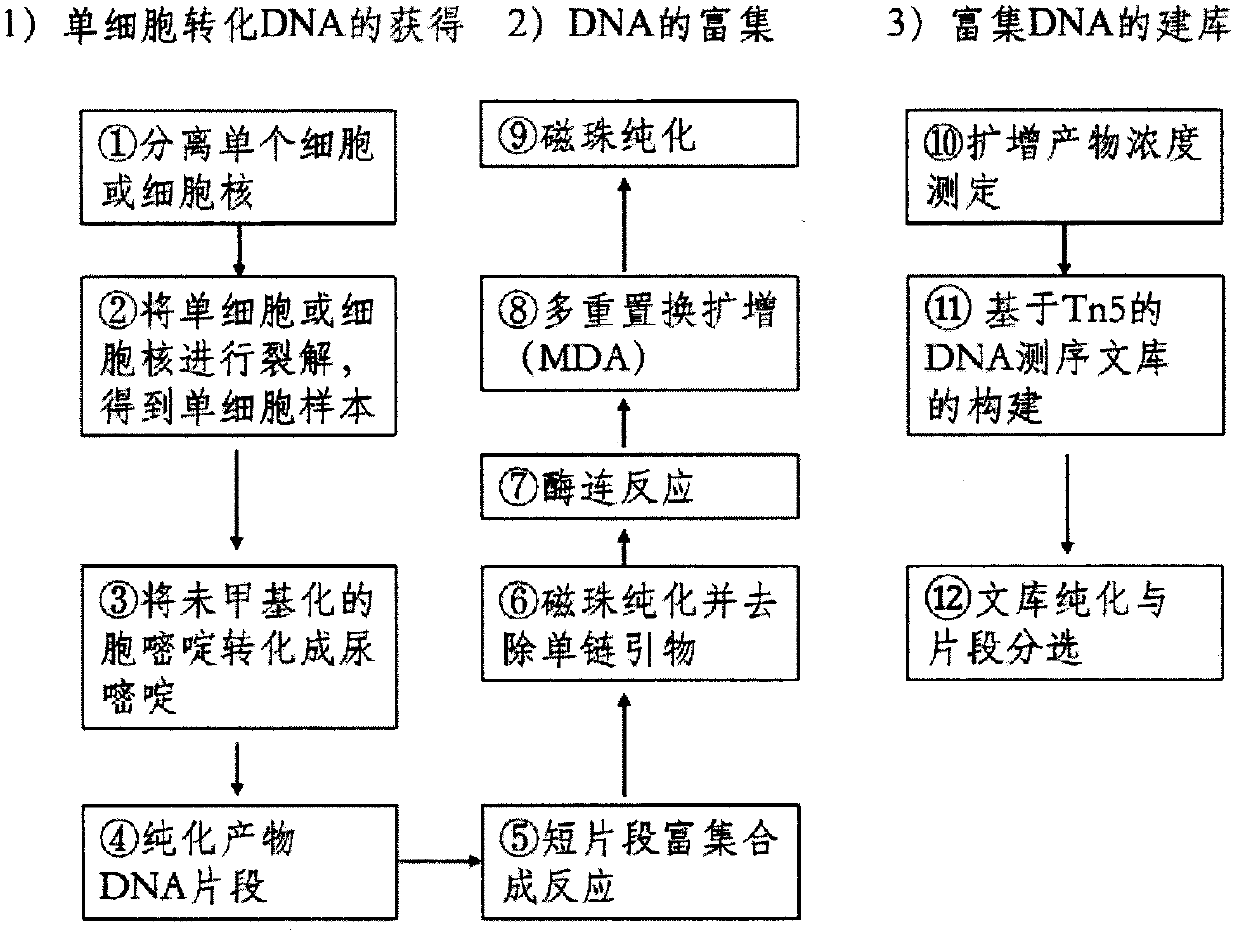
[0025] The operation process of the MB-seq sequencing method adopted by the present invention is as attached figure 1 As shown, the specific operation process is as follows:
[0026] 1. Put the tetrad microspores of corn into 27% mannitol solution, and the intracellular osmotic pressure balance maintains the normal cell shape. Using micro-glass tubes, micro-injectors, and microscopes, isolate single cells or nuclei in 4 μl of pure water. After separation, place the samples on ice until all samples are separated and proceed to the next step or move to a -80°C refrigerator for later use;
[0027] 2. Prepare 8μl cell lysis buffer (15mM Tris-Cl pH 7.4, 0.9% SDS and 0.5μl proteinase K), add the buffer to 4μl sample, incubate at 37°C for 1 hour, after the reaction, you can perform the next step or move to - Standby at 20°C;
[0028]3. Use the Imprint DNA Modification Kit to convert unmethylated cytosine to uracil:
[0029] a) Add 1 μl Balance Solution to the cleaved DNA above, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com