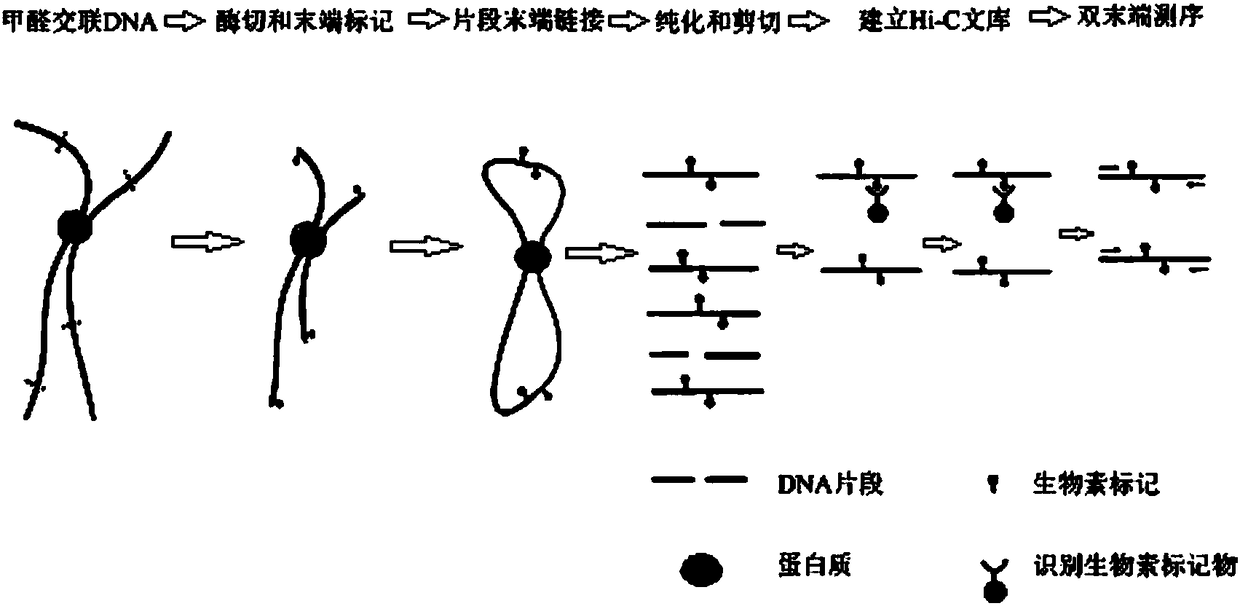
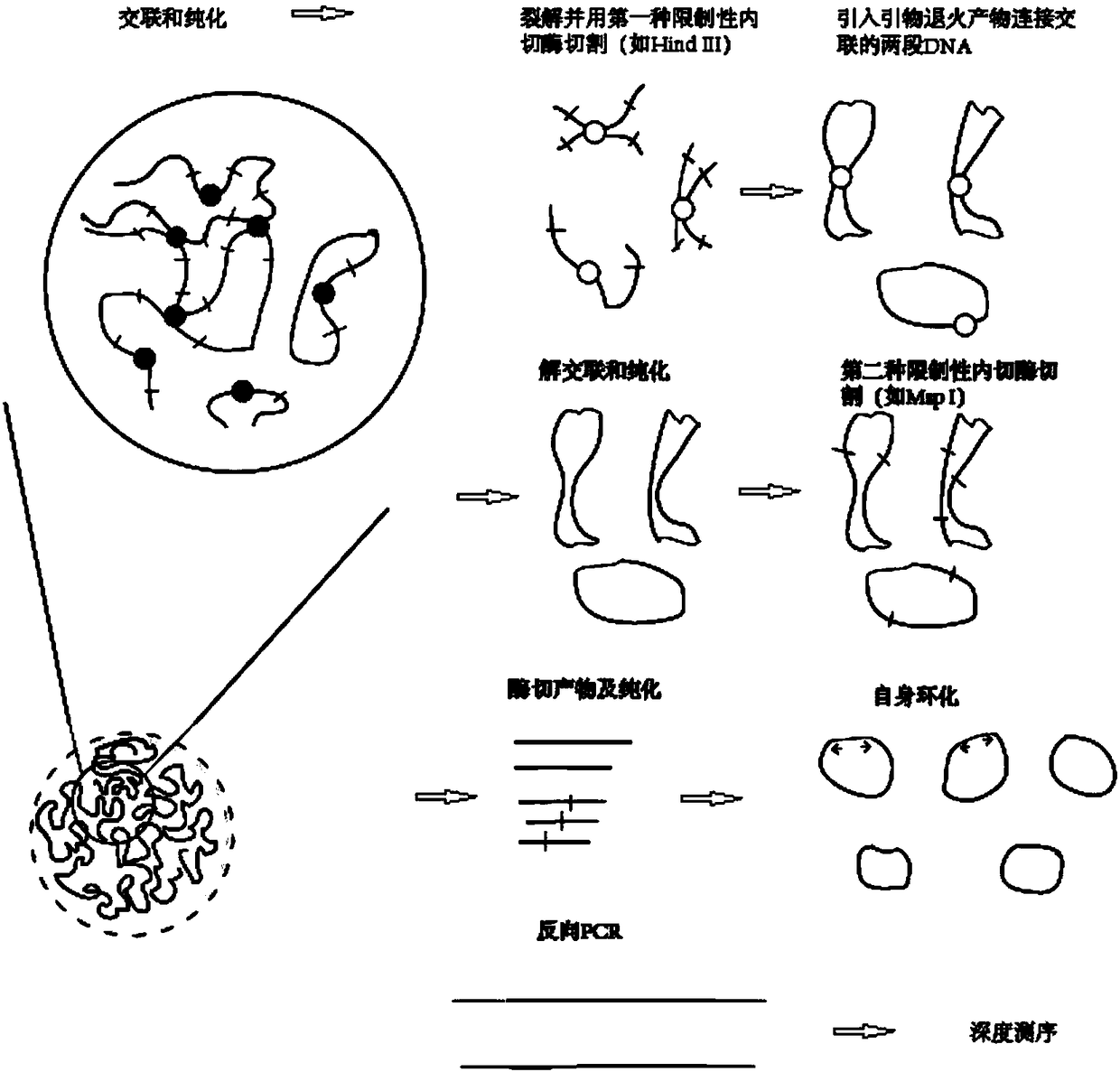
Whole genome interaction library and construction method thereof
A construction method and a genome-wide technology, applied in the field of high-throughput sequencing library construction, can solve cumbersome and time-consuming problems, and achieve the effect of wide operability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Choose 2g fresh leaves (approximately 25×10 6 Cells) were used as the starting material, and formaldehyde with a mass concentration of 1% was cross-linked at room temperature for 40-60 minutes to obtain cells with immobilized chromatin. The specific library construction steps were as follows:
[0039] 1) Cell lysis. Cells per copy (25×10 6 1) Add 1 ml of ice-cold lysate (10 mM Tris-HCL, 30 mM NaCl, 0.2% NP-40, 10% protease inhibitor, pH 7.4), and place on ice for 15 min. Submerge 4 groups of cells with a tissue homogenizer on ice, 30 times each. The lysate was centrifuged at 2,000xg for 5 min at room temperature. The samples were washed twice with pre-cooled restriction endonuclease corresponding digestion buffer (1×NEBuffer Cutsmart) and collected by centrifugation at 2000×g for 5 min. Resuspend the sample with 260 μL of this digestion buffer.
[0040] 2) Chromatin digestion. Add 1560 μL restriction endonuclease corresponding digestion buffer (1×NEBuffer Cutsmart...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com