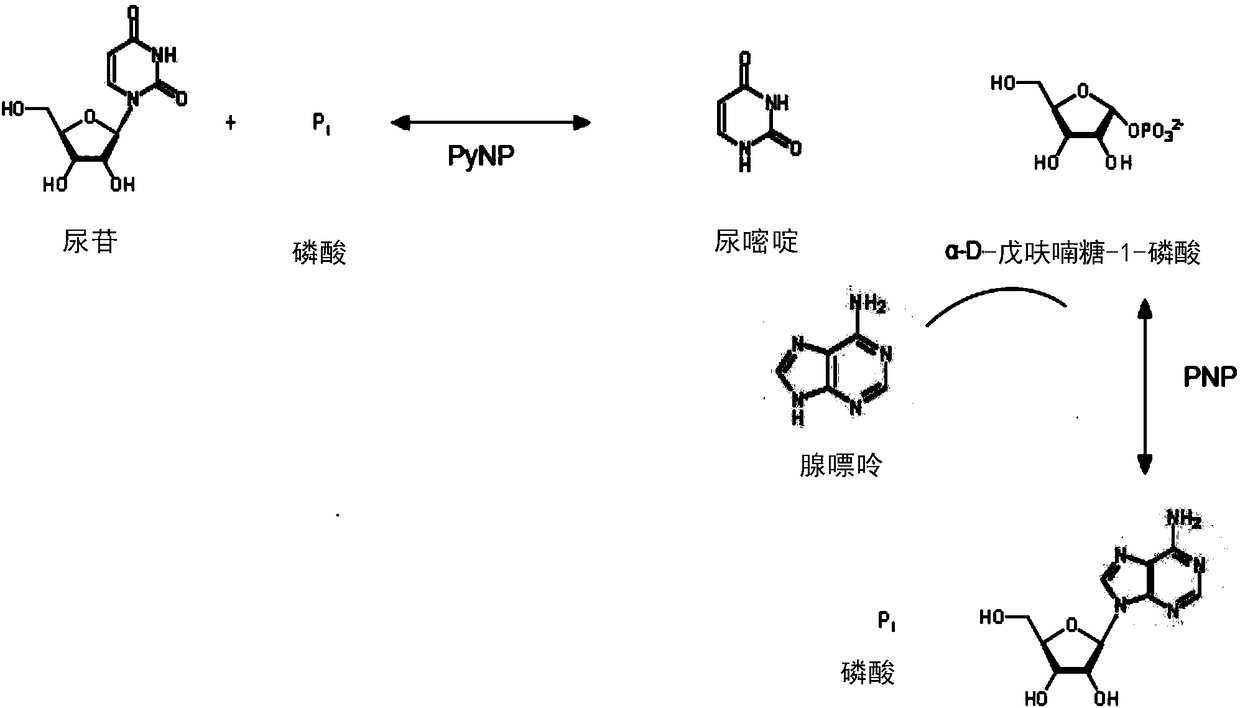
2'-deoxy-2'-fluoro-beta-D-arabinofuranosyladenine production method
A technology of glycoadenylic acid and glycouridylic acid, which can be used in biochemical equipment and methods, botanical equipment and methods, glycosyltransferases, etc., and can solve the problems of low conversion rate and limited large-scale application.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Embodiment 1, wild-type PNP, the construction of PyNP recombinant genetic engineering bacteria, expression and purification of recombinant protein
[0093] (1) Construction of recombinant plasmids
[0094] In this example, the wild-type PNP and PyNP genes and amino acid sequences used were derived from Escherichia coli. GenBank accession number M60917.2 for wild-type PNP; accession number U00096.3 for wild-type PyNP. Primers were designed and synthesized according to the sequences published in Genbank, and amplified by PCR.
[0095] PNP upstream primer sequence:
[0096] 5'-GGAATTCCATATGGCTACCCCACACATTAA-3' (SEQ ID NO: 5);
[0097] PNP downstream primer sequence:
[0098] 5'-GGCGAGCTCTTACTCTTTTATCGCCCAGCAG-3' (SEQ ID NO: 6);
[0099] PyNP upstream primer sequence:
[0100] 5'-GGAATTCCATATGCTTCAAAGTAATGAGTAC-3' (SEQ ID NO: 7);
[0101] PyNP downstream primer sequence:
[0102] 5'-GGCGAGCTCTTACAGATAGCGGCACAGATA-3' (SEQ ID NO: 8).
[0103] The reaction system of P...
Embodiment 2
[0116] The preparation of embodiment 2, PNP, PyNP mutant
[0117] Taking PNP as an example to make a statement, PyNP is consistent with it.
[0118] 1) Construction of mutant library
[0119] Using the recombinant pET-PNP-wt as the DNA template, in which the primers are T7 universal primers, a random mutant library was constructed by error-prone PCR, and by adjusting the concentrations of Mg2+ and Mn2+ in the error-prone PCR reaction system and dCTP and dTTP oligonuclear nucleotide concentration, so that the base mismatch rate of the mutant library is 5 / 1000, that is, to ensure that a mutant has 1 to 3 amino acid mutations, the specific process of constructing the mutant library is as follows:
[0120]
[0121]
[0122] T7 promoter primer: 5'-TAATACGACTCACTATAGGG-3' (SEQ ID NO: 9);
[0123] T7 terminator primer: 5'-GCTAGTTATTGCTCAGCGG-3' (SEQ ID NO: 10).
[0124] Reaction conditions:
[0125]
[0126] The obtained error-prone PCR product was subjected to electroph...
Embodiment 3
[0138] Example 3, directional mutation of wild-type PNP, PyNP, and purification
[0139] 1) Site-directed mutation of wild-type PNP and PyNP
[0140] Starting from PNP-wt and PyNP-wt, use QuikChange TM The Site-Directed Mutagenesis Kit performs site-directed mutagenesis according to the operation steps of the kit. The mutation of I at position 86 of PNP-wt to R is designated as "PNP-mutant"; the mutation of T at position 36 of PyNP-wt to R is designated as "PyNP-mutant".
[0141] 2) Shake flask fermentation and Ni column purification of PNP-mutant and PyNP-mutant
[0142] The specific steps are referring to embodiment 1.
[0143] 3) Carry out enzyme reaction with the purified enzyme, the enzyme reaction conditions are (final concentration): 2mM 2'-FANA-U, 3mM adenine (A), 1mM pH7.4 phosphate buffer solution, PNP mutant enzyme 10ul / ml, PyNP mutant enzyme 10ul / ml, react at 37°C for 48h,
[0144] HPLC measures the conversion rate, as shown in Table 3 and Table 4.
[0145] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com