Cell elution method based on drug target residence time
A residence time, cell technology, applied in the field of cell elution, which can solve the problems of low screening throughput, inconvenient operation, and dependence on radioisotope labeling ligands.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
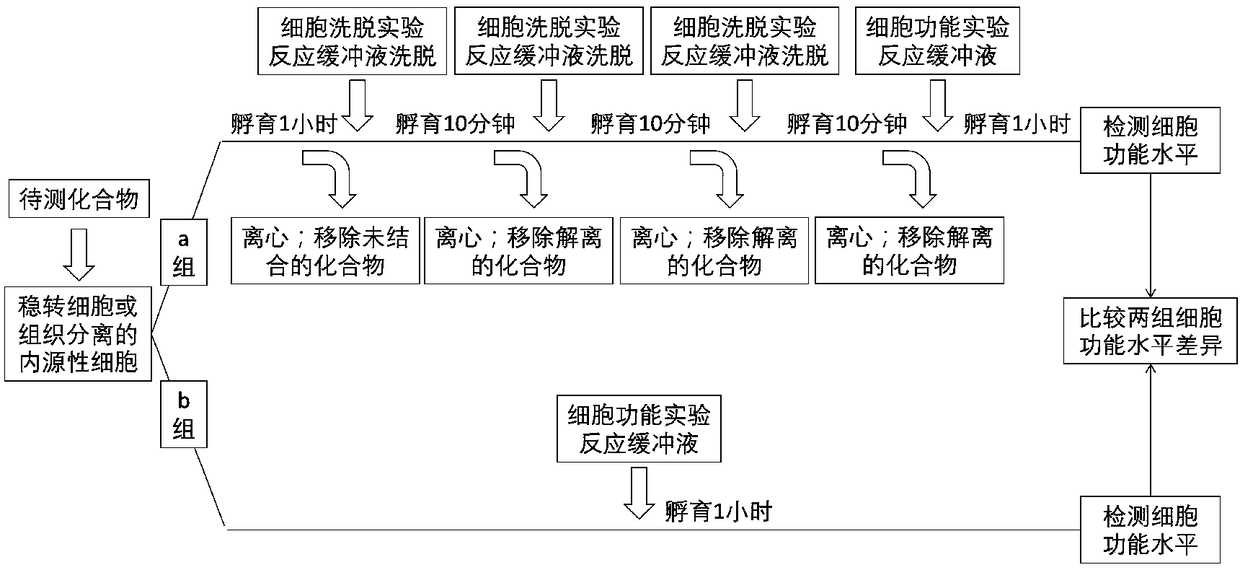
[0012] A cell elution method based on the residence time of the drug target, the method is: using highly expressed human adenosine A 1 CHO / ADORA1 stably transfected cells for the receptor screen for adenosine A with different residence times 1 Receptor agonists, the specific steps are as follows:
[0013] (1) CHO / ADORA1 cells were cultured and collected in vitro, divided into two equal parts, marked as group a and group b, each containing 4000 cells;
[0014] (2) Add 0.8U / mL adenosine deaminase to the DMEM / F12 medium as the reaction buffer for cell elution experiments; place the cells in group a in the prepared reaction buffer for cell elution experiments; add excess The compound to be tested was incubated at 37°C for 1 hour to allow the compound to be tested to interact with the adenosine A expressed on the cells 1 The receptors are fully bound; after incubation for 1 hour, transfer the above cell mixture to a centrifuge tube, centrifuge at 4°C for 3 minutes at a speed of 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com