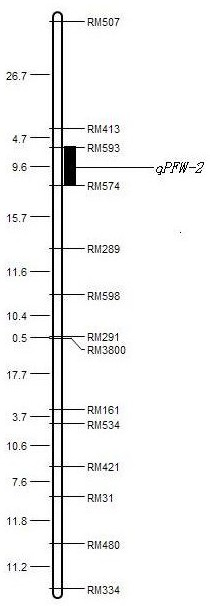
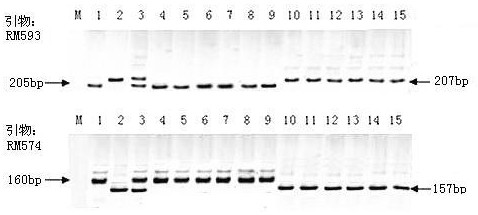
Mining and application of SSR markers closely linked to rice salt-tolerance QTL on chromosome 5
A number chromosome, chromosome technology, applied in the field of agricultural science, can solve problems such as difficulty, complex genetic basis, slow progress, etc., to achieve the effect of speeding up the process, clear site location, and convenient identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
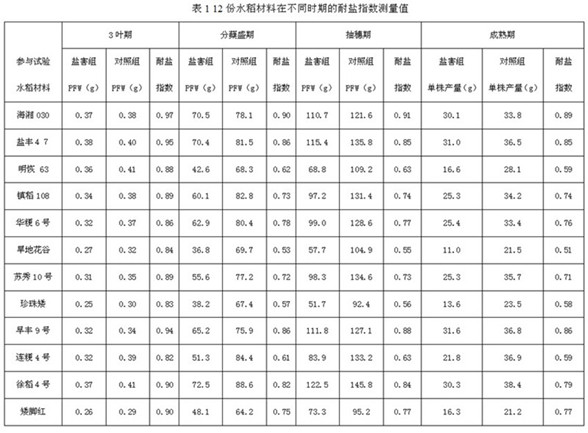
[0030] At the beginning of April 2015, 12 rice varieties with obvious differences in salt tolerance were selected for many years of field planting to carry out the identification of salt damage in rice ponds in the greenhouse of Dongxin Farm. The fresh weight of the plant and the yield per plant at the ripening stage were used to obtain the salt tolerance index of 12 rice varieties.
[0031] The methods for identifying salt damage in rice ponds are as follows: (1) Salt pond preparation: use stainless steel to build the salt pond. The salt pond is a long groove with a height of 35cm, a width of 35cm, and a length of 2.5m. The non-salinized soil was crushed, sieved, and then dried in the sun. An equivalent amount of 200 kg of non-salinized soil was added to each salt pool, and the salt damage treatment group and the control group were set up respectively. The salt damage treatment group used 80 kg of 0.5% NaCl salt solution. Soil soaking, the control group used the same amount o...
Embodiment 2
[0037] In the first season of 2015, the excellent salt-tolerant rice variety Haixiang 030 was selected as the female parent, and the salt-sensitive variety Lianjing No. 4 was selected as the male parent, and hybrid F1 was obtained. The F2 segregation population, the F2:3 family was obtained by selfing of the F2 single plant in the 2016 season.
[0038] In May 2017, the rice pond salt damage identification method was adopted for the F2:3 family. The detection index selected the plant fresh weight (plant fresh weight, PFW) at the peak tillering stage of rice. Ben's salt tolerance index.
[0039] The leaves of Haixiang 030×Lianjing 4 parent, F1 and F2:3 mapping populations were taken, and the DNA was extracted by SDS crude extraction method, and 368 pairs of SSR primers that were roughly evenly distributed on the 12 chromosomes of rice in the laboratory were used. Parents, F1 and F2:3 mapping populations were amplified by PCR (the PCR reaction system for PCR amplification was: t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com