Recombinant Clostridium beijerinckii for efficient sucrose fermentation and method for improving sucrose fermentation performance of Clostridium beijerinckii
A technology of Clostridium beijerinckii and sucrose, which is applied in the field of genetic engineering, can solve the problem that the performance of Clostridium beijerinckii for fermentation and utilization of carbon sources cannot be accurately controlled, etc., to improve the transport efficiency of sucrose, increase the yield of butanol, and improve the high-efficiency utilization Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Construction of the recombinant expression plasmid pSOS95-SUT1 containing the sucrose transporter gene SUT1
[0048] Amplify the linearized pSOS95 vector portion by PCR:
[0049] Upstream primer: 5'-TAAAAATAAGAGTTACCTTAAATGGTAACT-3', (SEQ ID NO.3);
[0050] Downstream primer: 5'-TTTAATCCCTCCTTTTTAAATTCTGGATCCT-3', (SEQ ID NO.4).
[0051] The PCR reaction program was: pre-denaturation at 98°C for 3 min, 30 cycles at 98°C for 10 s, 60°C for 15 s, 72°C for 5 min, and finally extension at 72°C for 10 min.
[0052] Using PCR technology to amplify the target gene SUT1:
[0053] Upstream primers:
[0054] 5'- CCAGAATTTAAAAGGAGGGATTAAA ATGGAGAATGGTACAAAAAGAGAAGG-3', (SEQ ID NO. 5);
[0055] Downstream primers:
[0056] 5'- CCATTTAAGGTAACTCTTATTTTTA TTATTTAATGGAAAGCCCCATGGCGAC-3', (SEQ ID NO. 6);
[0057] Underlined parts in italics are homologous sequences to the vector.
[0058] The PCR reaction program was: 98°C pre-denaturation for 3 minutes, 98°C for 1...
Embodiment 2
[0060] Example 2: Construction of recombinant Clostridium beijerinckii 8052-SUT1 expressing SUT1 gene heterologously
[0061] The expression plasmid pSOS95-SUT1 containing the sucrose transporter gene and the empty vector plasmid pSOS95K were respectively transferred into E.coli JM109 (pAN1) for methylation, and the methylated plasmids pSOS95-SUT1 and pSOS95K were respectively introduced by electroporation In C. beijerinckii 8052, positive transformants were screened on a TYA plate containing 50ug / mL erythromycin.
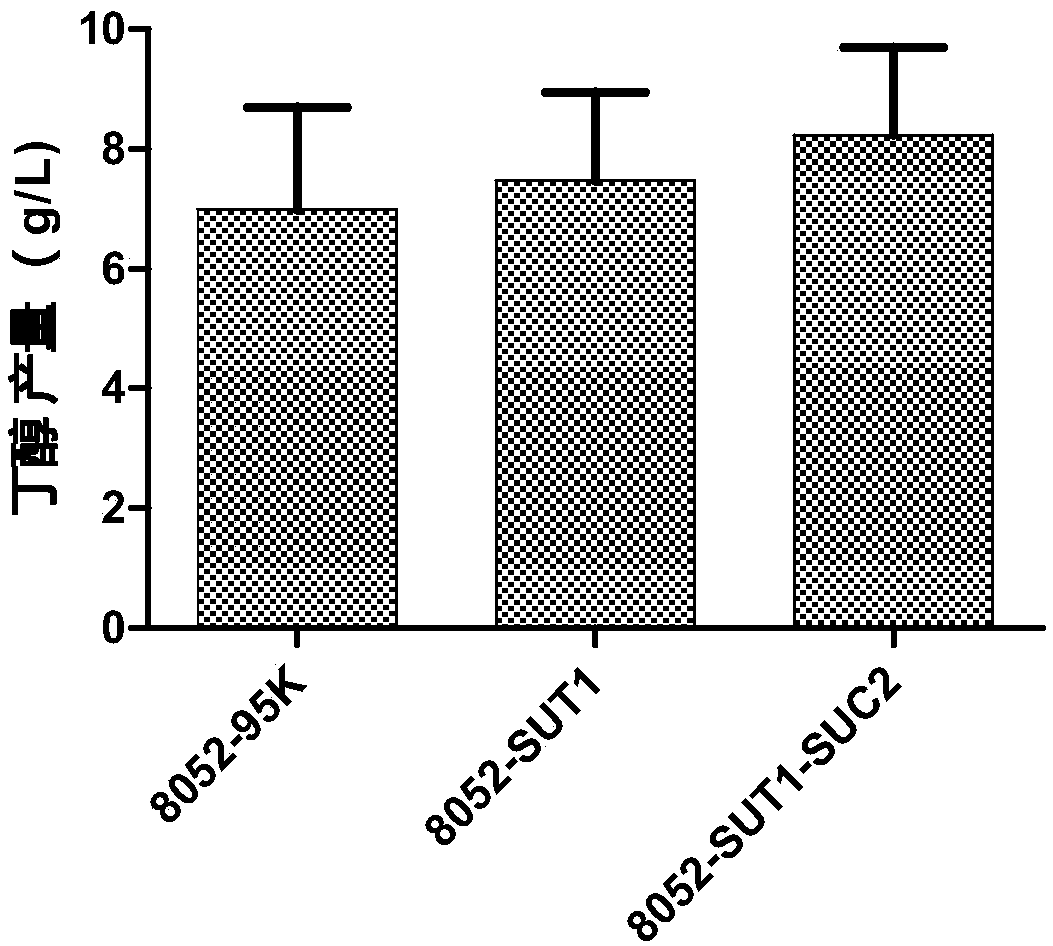
[0062] The culture temperature is 30°C, and the culture time is 2-3 days. Select transformants from the selection plate, and obtain positive recombinant strains after colony PCR verification (upstream primer: AGGCATTAGTGCATTTAAGC (SEQ ID NO.7), downstream primer: CCAGGCTTTACACTTTATGC (SEQ ID NO.8); PCR reaction program: 98 ° C pre-denaturation 10min, 98°C for 10s, 53°C for 15s, 72°C for 2min, 30 cycles, and finally 72°C for 10min). figure 2 .
[0063] The recom...
Embodiment 3
[0064] Example 3: Construction of recombinant expression plasmid pSOS95-SUT1-SUC2 containing sucrose transporter gene SUT1 and sucrase gene SUC2
[0065] PCR amplification of the linearized pSOS95-SUT vector portion:
[0066] Upstream primer: 5'-TAAAAATAAGAGTTACCTTAAATGG-3', (SEQ ID NO.9);
[0067] Downstream primer: 5'- CCCTCCT TTATTTAATGGAAAGCCCCATGGCGACTGC-3', (SEQ ID NO. 10).
[0068] (The part in italics underlined in the primer is the SD sequence)
[0069] The PCR reaction program was: pre-denaturation at 98°C for 3min, 30 cycles at 98°C for 10s, 57°C for 15s, 72°C for 6min, and finally extension at 72°C for 10min.
[0070] PCR amplification of the SUC2 gene:
[0071] Upstream primer: 5'-
[0072] GGCTTTCCATTAAATAA AGGAGGGATTAAAATGACAAACGAAACTAGCGATAG-3', (SEQ ID NO. 11);
[0073] Downstream primers:
[0074] 5'- CCATTTAAGGTAACTCTTATTTTTA CTATTTTACTTCCCTTACTTGG-3', (SEQ ID NO. 12).
[0075] (The underlined italic part in the primer is the homologous sequence...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com