SNP locus relevant to rapid growth of crassostrea gigas
A long oyster, site technology, applied in the field of genetic breeding, can solve the problems of slow growth, long oyster germplasm degradation, red tide, etc., achieve the effect of fast growth and reduce blindness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1: Acquisition of markers associated with rapid growth of the long oyster
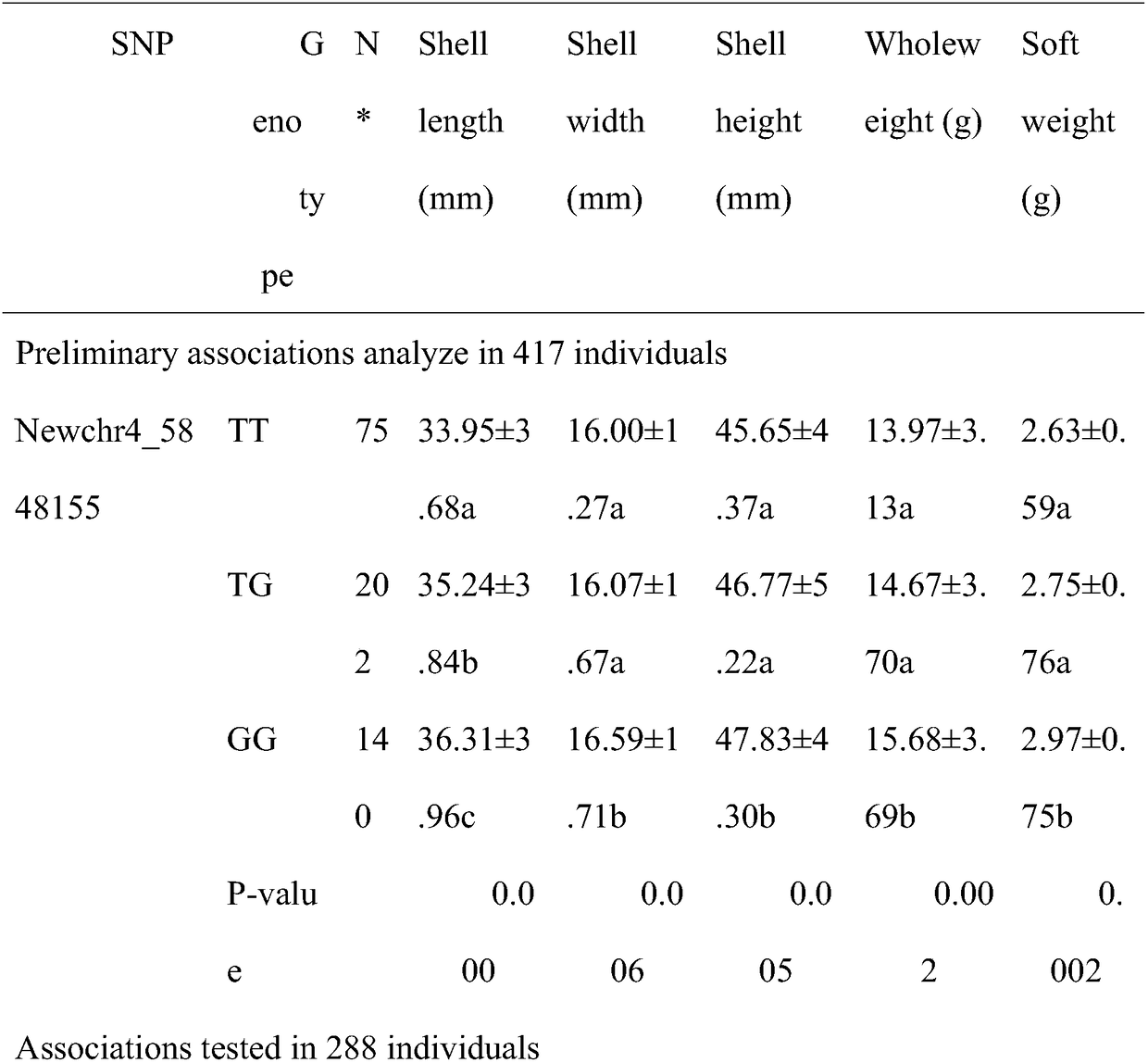
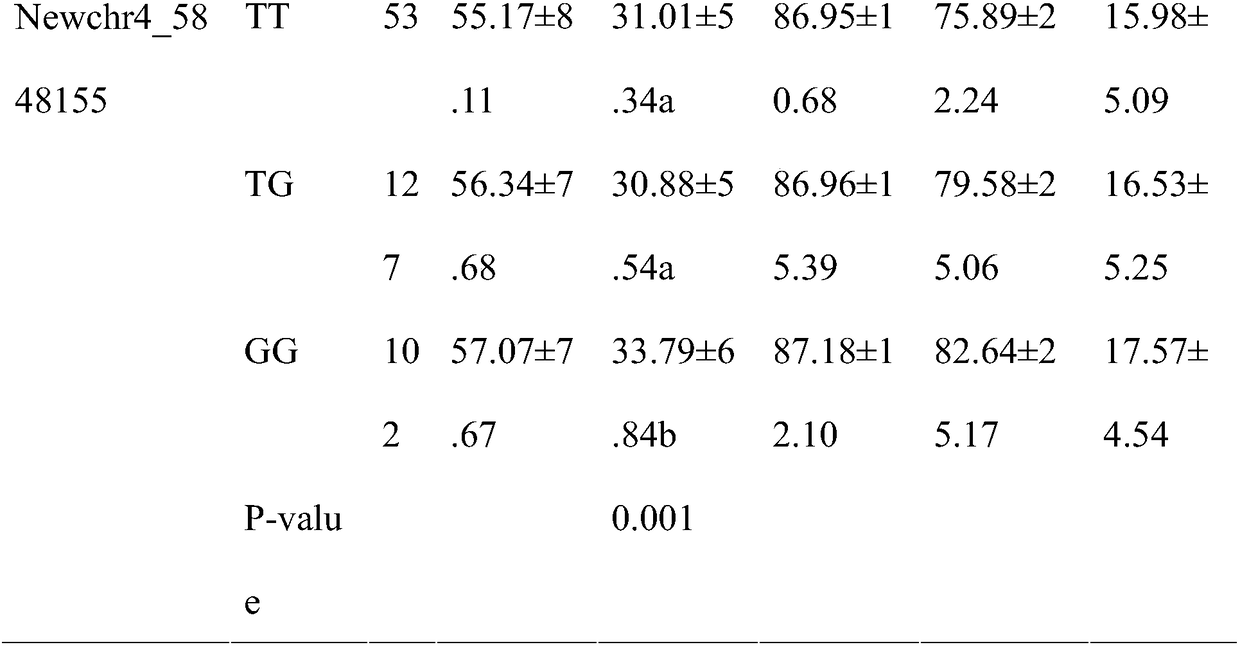
[0018] 1. On the basis of the complete sequencing of the long oyster genome, the applicant obtained the growth-related genes—bHLH family through GWAS association analysis technology. The bHLH family of oyster oyster was classified, including 89 genes from groups A to F. On this basis, 5 candidate genes were selected for SNP screening by resequencing, and a wild population of 417 individuals was selected. The obtained 63 loci were verified by gene chip method in the wild population of 288 individuals, and finally the correlation analysis with growth traits was carried out with biological software to obtain the NewChr4_588155SNP marker. The GG shell width of NewChr4_588155-marked genotype individuals is significantly larger than that of TT genotype and TG genotype individuals, and it is located in the exon of gene LOB (SEQ ID NO: 1), which changes the composition of the amino acid sequence...
Embodiment 2
[0026] Example 2: Application of SNP markers in screening fast-growing long oyster parents
[0027] A kit for evaluating and screening the SNP markers of fast-growing long oyster parents was made to detect the SNP site NewChr4_588155 of the LOB gene.
[0028] The kit contains reagents for specific amplification primers and specific extension primers required to detect SNP sites, wherein the specific amplification primer sequences for site NewChr4_588155 are SEQ ID NO: 2 and SEQ ID NO: 3, the The kit also includes common reagents required by corresponding techniques such as (10×PCR (containing Mg2+) buffer, dNTP (10mM), double distilled water, enzymes, etc. In addition, there may be standard products and controls such as determined genes).
[0029] The amplification reaction system includes: 1.0 μL of cDNA, 2.5 μL of 10×PCR (containing Mg2+) buffer, 2.0 μL of dNTP with a concentration of 10 mM, 1.0 μL of an upstream primer with a concentration of 10 μM, 1.0 μL of a downstream p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com