InDel primer identifying late-bolting QTL located on radish R02 chromosome and application of primer
A chromosome and indel520 technology, applied in the biological field, can solve the problems of insufficient linkage distance, different, difficult to apply, etc., and achieve the effect of saving the cost and time of plant field management
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
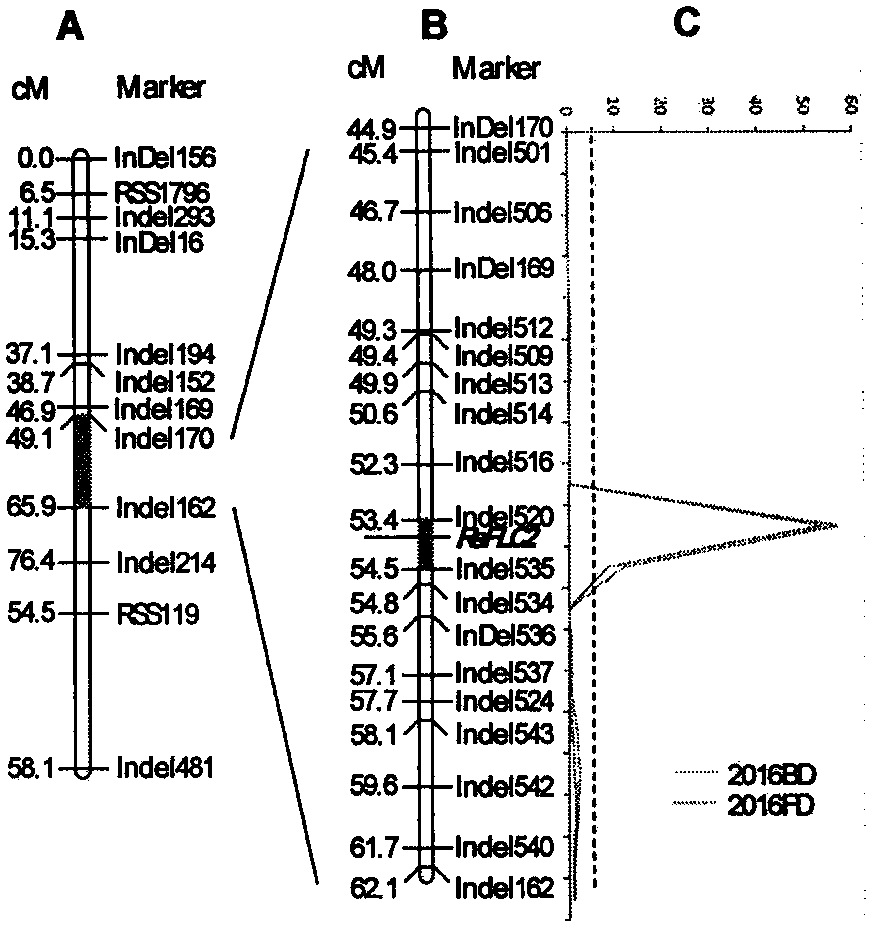
[0060] Acquisition of Molecular Markers for Late Bolting Trait in Radish
[0061] The invention utilizes a conventional map-position cloning method to identify and clone genes related to late bolting traits of radish.
[0062] 1. Genetic segregation population construction and phenotypic identification
[0063] Using the Japanese late bolting material "Ninengo" and the Chinese early bolting material "Maer" as parents, a F 2 group. Parents, F in the spring of 2016 1 , F 2 The bolting and flowering traits of individual plants in the population were investigated. The survey method is: survey the budding and flowering of individual plants every other day. Budding time is the number of days required from planting to visible flower buds; flowering time is the number of days required from planting to the opening of the first flower. The budding time and flowering time of the late bolting material "Ninengo" were 91 days and 110 days, respectively, and the budding time and flower...
Embodiment 2
[0073] Application of InDel520 and InDel535 in Identification of Radish Late Bolting and Early Bolting Materials
[0074] 1. Identification of late-bolting and early-bolting radish materials by molecular markers
[0075] 1) DNA extraction
[0076] Genomic DNA of 183 kinds of radish materials to be tested in Table 2 were extracted by conventional CTAB method. The 183 tested radish materials in Table 2 are the F1 generation obtained by crossing the parents Ninengo and Maer, and the F2 individual plants obtained by selfing the F1.
[0077] 2) PCR amplification and detection
[0078] The above-mentioned InDel520 primer pair and InDel535 primer pair were used for PCR amplification of the radish material to be tested.
[0079] PCR reaction system: containing 100ng genomic DNA, 1μl 10×PCR Buffer, 0.8μl dNTPs, 0.2μl (10μM) of the above-mentioned upstream and downstream primers, 1U Taq enzyme, plus ddH 2 0 to 10 μl. The PCR amplification program was: pre-denaturation at 94°C for 4...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com