Intelligent leukemia cancer cell detection instrument
A technology for leukemia and cancer cells, applied in the application field of gene microarray data, can solve the problem of finding the best parameters for the optimal feature subset classification of gene microarray data, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
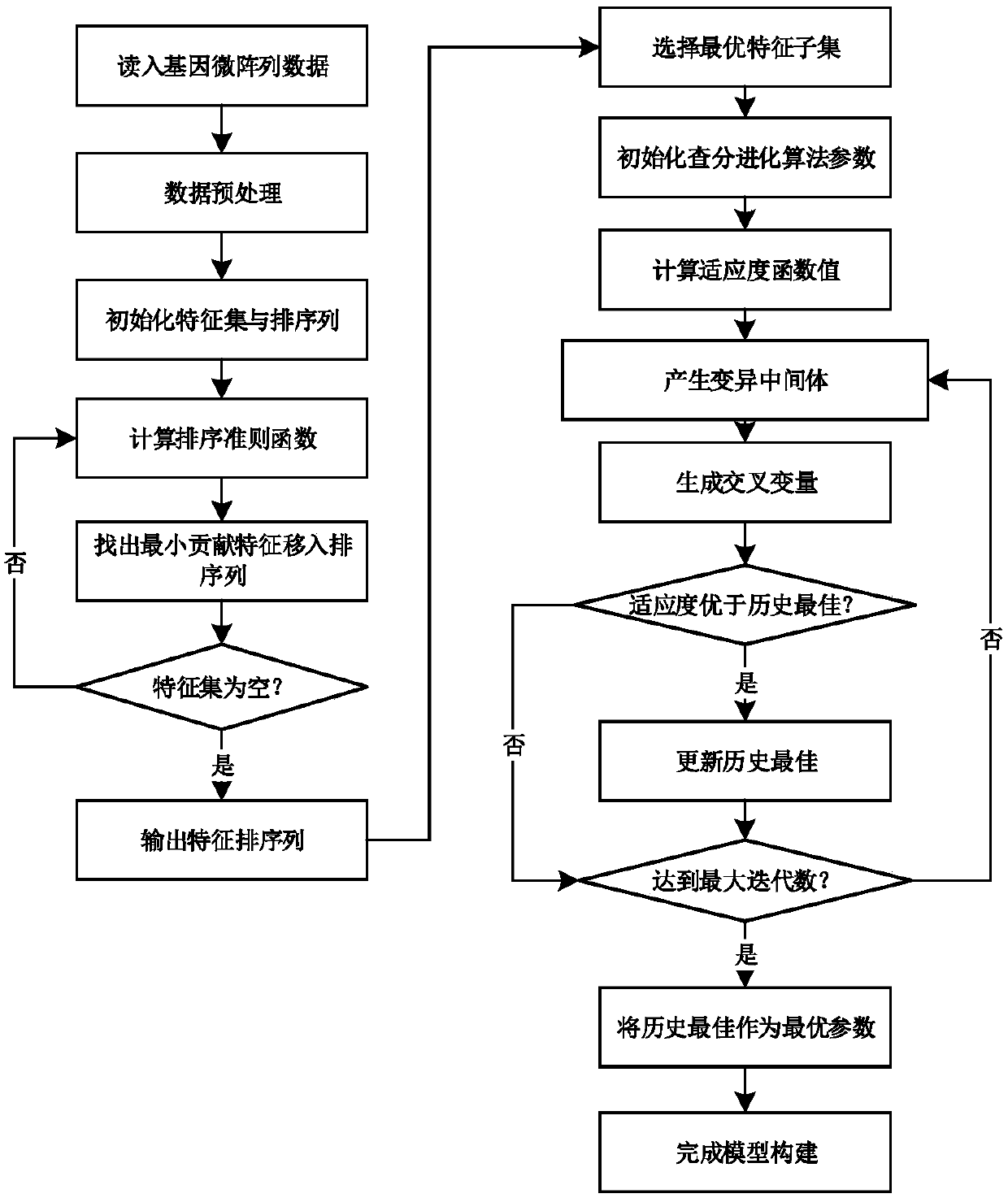
[0050] The present invention will be described in detail below according to the accompanying drawings.
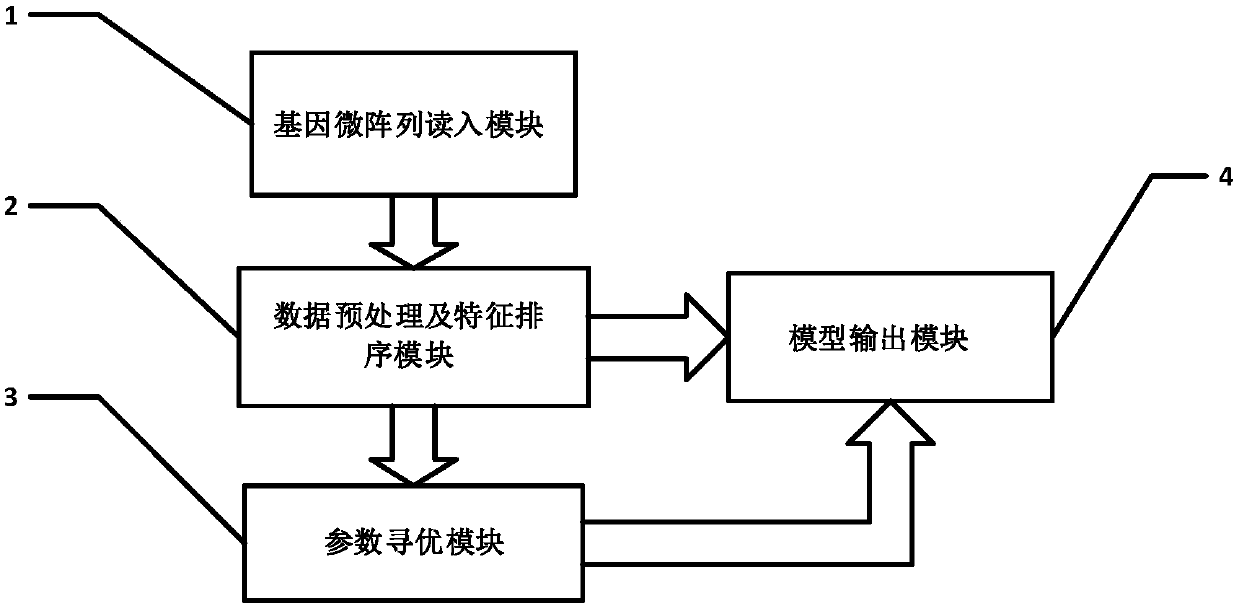
[0051] refer to figure 1 , an intelligent leukemia cancer cell detector, the system consists of a gene microarray read-in module 1, a data preprocessing and feature sorting module 2, a parameter optimization module 3, and a model output module 4; wherein:
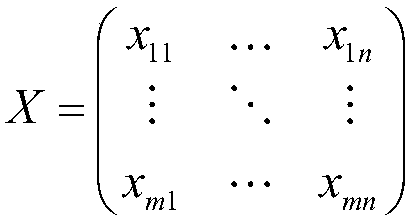
[0052] Gene microarray read-in module 1, which reads in the category labels of all gene microarrays Y=[y 1 ,y 2 ,...,y m ], where y i =k,k∈(-1,1), and the gene microarray expression values of all samples:
[0053]
[0054] where each line x i Represents the expression values of all genes in a sample, corresponding to each column x j Represents the expression value of a gene in all samples, the subscript i indicates the i-th sample, a total of m, and the subscript j indicates the j-th gene, a total of n.
[0055] The data preprocessing and feature sorting module 2 is the process of normalizing and feature so...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com