Screening method and application of target gene SSR molecular markers of acanthopanax senticosus
A screening method and molecular marker technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., to speed up the breeding process and overcome uncertainties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
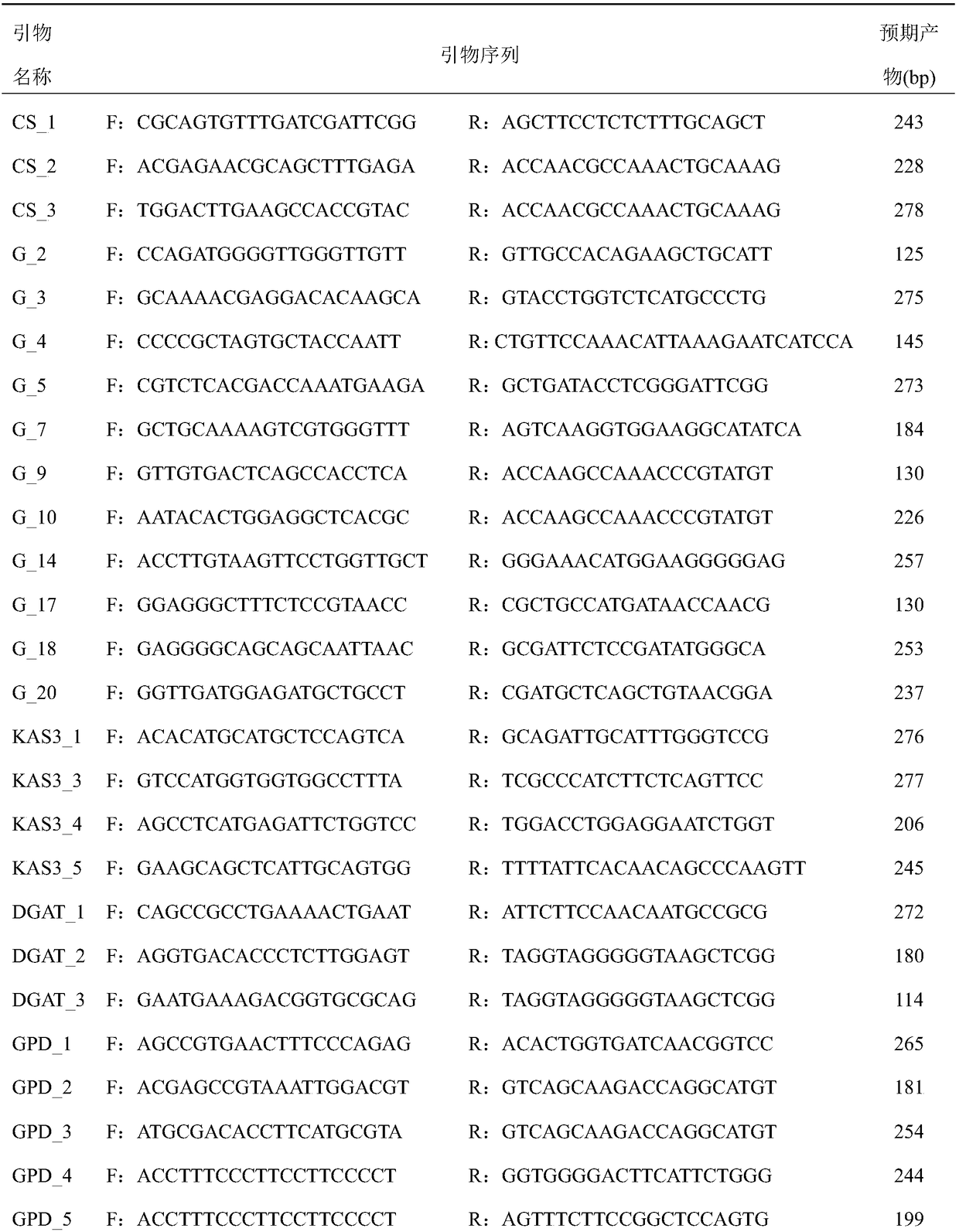
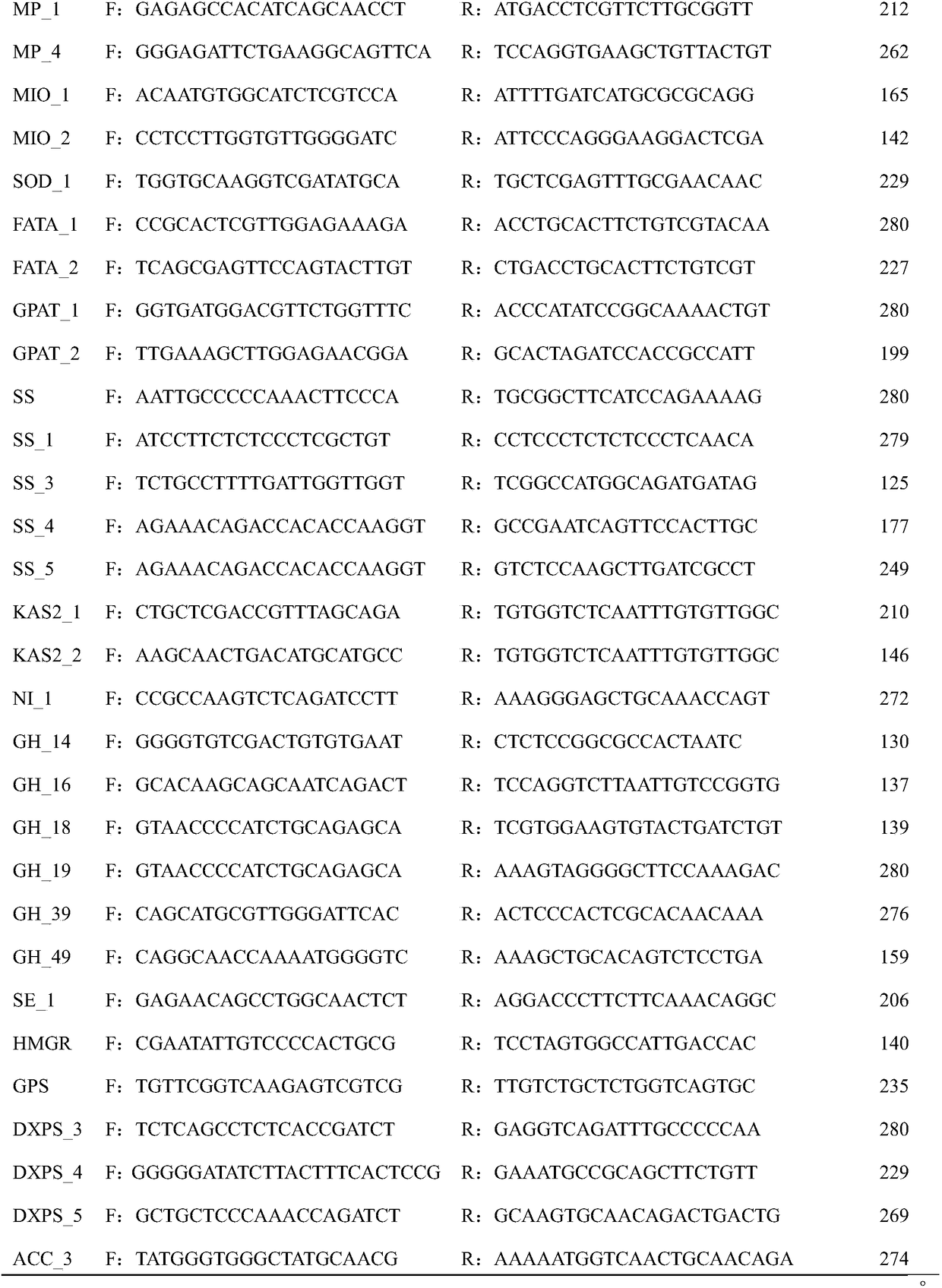
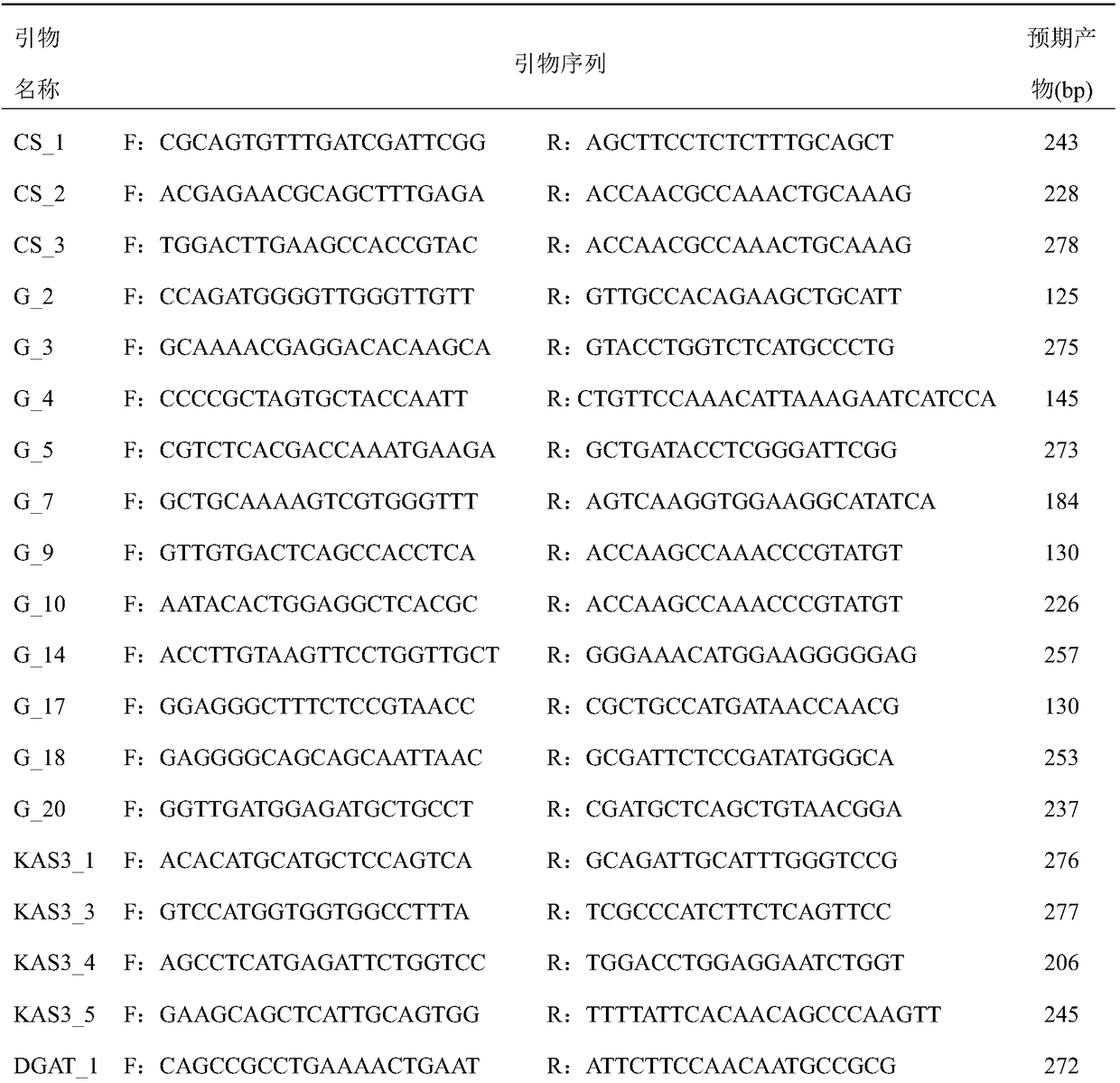
[0017] 1. Extract total RNA from Acanthopanax fruit, root, stem and leaf
[0018] One Acanthopanax germplasm SL1 from Suileng County, Heilongjiang Province was used as material. The total RNA of Acanthopanax senticosus from different tissues and organs was extracted according to the method recommended by Shanghai Shenggong Plant RNA Extraction Kit. The total RNA concentration of each tissue and organ was 200-400ng / μL, and stored at -80°C.
[0019] 2. RNA-seq data analysis of Acanthopanax senticosus fruit, root, stem and leaf
[0020] A transcriptome library was constructed by reverse transcription using samples mixed with equal amounts of total RNA from four tissues and organs as templates. Clean reads were obtained by removing adapters, N ratio greater than 0.1% and low-quality reads from the raw reads obtained by sequencing, and the clean reads were spliced using Trinity software to obtain unigene. Compare unigenes with known functional genes in public databases (Nr, Sw...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com