Application of NW_006882077-1 in CHO cell genome to stable expression of protein
A stable expression and cell gene technology, applied in the field of genes, can solve the problems of time-consuming and laborious screening of high-expression monoclonal, hinder the efficiency of recombinant cell lines, and high cost, so as to avoid high-expression monoclonal screening and overcome the uncertainty of integration sites performance and reduce development time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
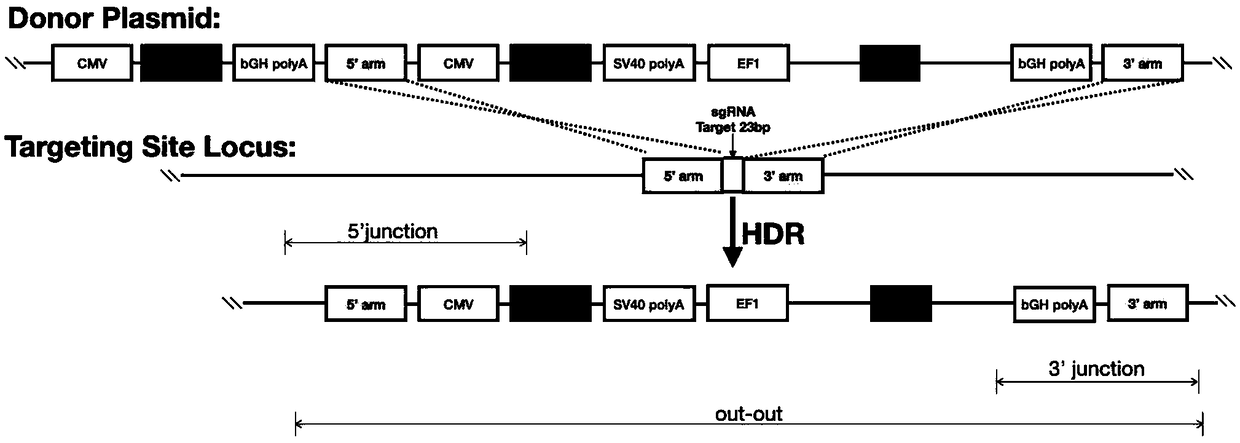
[0042] Selection of high expression sites;
[0043] NW_006882077.1 was integrated at base 691045, where the Zsgreen1 gene was integrated. For this fluorescent cell, subculture was carried out for no less than 50 generations, and the expression level of its fluorescence was detected by flow cytometry. The 50th generation of fluorescent cells still has a good expression level of green fluorescent protein, and the fluorescent signal can be stably retained during the passage of cells.
[0044]In addition, this fluorescent cell was also subjected to suspension acclimation, and the expression level of the fluorescent protein after suspension acclimation was detected again by flow cytometry. The test results show that more than 95% of the recombinant CHO cells that have been suspended for 50 passages still maintain the expression of green fluorescent protein after suspension. It can be considered that this site is extremely stable and will not be lost due to cell passage. fluoresce...
Embodiment 2
[0046] selection of specific targets;
[0047] According to the principle of proximity, the sequence:
[0048] 5'CCAATGCTATTACCTTCTTCAGCATCTGTTCTTGAAGACCTTCAGTCACTTTAGTTAACTCTTTCCACTTTCGTTACTGCAACTCTCAGATC-TAACTTGGCTTG 3'
[0049] Input into the CRISRPRater system to predict and select target sequences with low off-target efficiency. The parameter settings are as follows: 1) The maximum number of mismatched bases in the first 15 bp after NGG is 0; 2) The number of mismatched bases in all 21 bp after NGG is 2.
[0050] After the above operation, the following sequence with a score of 0.87 was selected as the target sequence according to its score:
[0051] 5'-GATCTAACTTGGCTTGCCTGAGG-3';
[0052] According to the CRISPRater system, LOW efficacy (score0.74).
[0053] According to the CRISPRater evaluation system, all the target sequences in the range of 690980-691090 near the site NW_006880285.1 obtained a score of more than 0.56, all of which were in the moderately effective...
Embodiment 3
[0054] The selection of embodiment 3 promoter
[0055] Replace the above CMV (strong mammalian expression promoter derived from human cytomegalovirus) promoter position with different promoters, including EF-1a (strong mammalian expression promoter derived from human elongation factor 1α), SV40 (simian Mammalian expression promoter derived from vacuolar virus 40), PGK1 (mammalian promoter derived from phosphoglycerate kinase gene), UBC (mammalian promoter derived from human ubiquitin C gene), human beta actin (β- Actin gene-derived mammalian promoter) or common promoters such as CAG (strong hybrid mammalian promoter). After detection, the above promoters can all regulate the downstream red fluorescent protein gene sequence, and express the corresponding red fluorescent protein.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com