Preparation method of citrus dominant function mutant
A mutant and functional technology, applied in the field of biotechnology research and development, can solve the problems of difficult plant materials and long citrus childhood, and achieve the effect of improving the infection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0043] The invention provides a method for preparing a citrus dominant functional mutant, comprising the following steps:
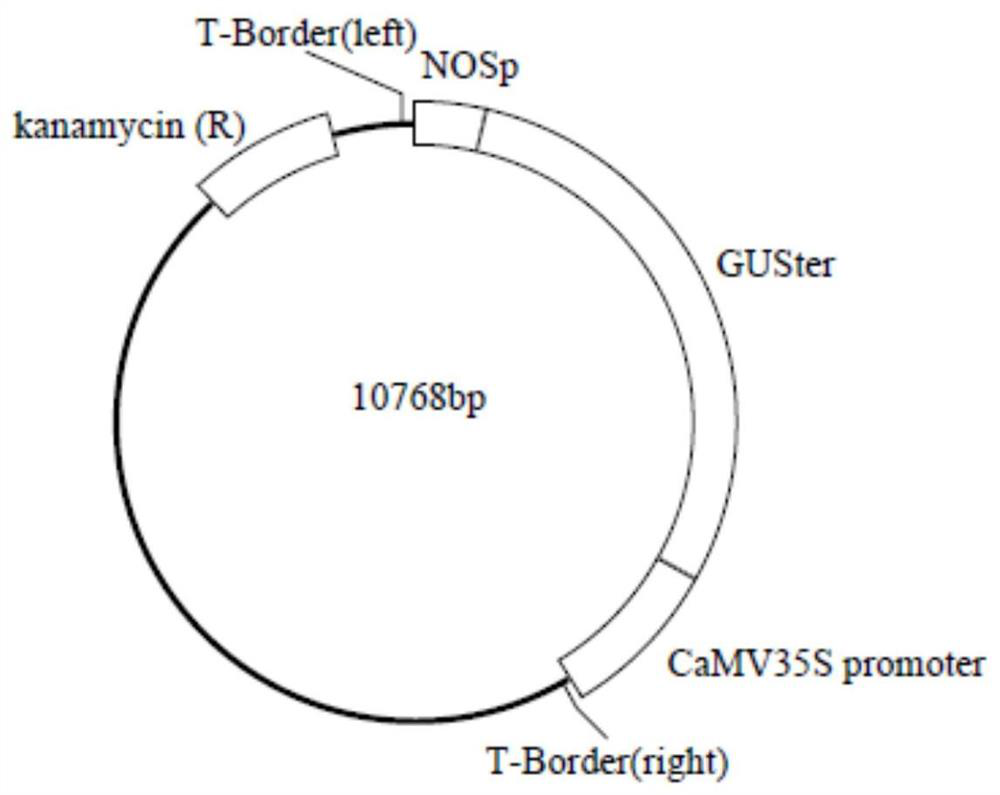
[0044] Step 1, using the first pair of primers shown in SEQ ID NO: 1 and 2, using the pCAMBIA1300 plasmid as a template, PCR amplification obtains the vector backbone containing the transfer DNA domain, and then the first strong promoter gene, reporter Both the gene and the second strong promoter gene are connected between the left and right boundaries of the transfer DNA domain of the vector backbone, and the destination vector containing the target sequence in the transfer DNA domain is constructed, wherein the first strong promoter is located in the reporter gene upstream of , the second strongest promoter is located downstream of the reporter gene. The vector backbone of the present invention contains a transfer DNA domain, and the distance between the right border and the second strong promoter is suitable, a distance of more than ten base pairs, so ...
Embodiment 1
[0074] 1.1 Obtaining carrier elements
[0075] a. Use primers Right298XS-F (SEQ ID NO: 1: 5'-GCGTCGACTCTAGAGGTAAACCTAAGAGAAAAGAGCG-3') and Left6582HS-R (SEQ ID NO: 2: 5'-GCGTCGACAAGCTTTTGTTTACACCCACAATATATCCTGC-3') to amplify plasmid pCAMBIA1300 to obtain vector element pVC ( 6286bp) PCR product. (The pCAMBIA1300 plasmid comes from the laboratory of Hunan Horticultural Research Institute; PCR reaction system (30 μL): 3 μL of 10×PCR buffer, 1 μL of dNTPs (each 2.5 mmol / L), 0.5 μL of forward primer (10 μmol / L), reverse Primer (10 μmol / L) 0.5 μL, Pyrobest DNA polymerase (5U / μL) 1 μL, template (0.2 μg / L) 1 μL, sterile deionized water 23 μL; amplification conditions: 95°C for 5min; 95°C for 20s, 55°C ℃20s, 72℃6min, 25 cycles; 72℃10min.)
[0076] b. with primers NOSp2462H-F (SEQ ID NO: 5'-CCAAGCTTGCCAATATCCTGTCAAACACTG-3') and NOSp2837S-R (SEQ ID NO: 6: 5'-GCGTCGACGCGAAACGATCCAGATCC-3') amplified plasmid pBI121 to obtain vector element NOSp ( 377bp) PCR product.
[0077] c. Usin...
Embodiment 2
[0113] 1. Preparation of dominant functional mutants of rock sugar orange and red navel orange
[0114] ① Seeds of rock sugar orange or navel orange, which were stripped of the outer testa and treated with 1% NaClO for 10 min, were sown on MS salt medium (pH 5.7) and cultured in the dark for about 20 days, and then cultured in the light (12 h light / 12 h dark) for 10 days. The growth temperature was maintained at 26-28°C. At this time, the thickness of the stem of the seedling plant reaches 1mm-3mm, and the plant height is greater than 5cm.
[0115] ② Pick a single colony of Agrobacterium (the strain contains pVC-NOSpGUSter-35 plasmid or pBI121 plasmid) in LB liquid medium (containing 50 μg / mL Rif and 50 μg / mL kanapenicillin), in a shaker at 28 ° C, Cultivate overnight at 180rpm; then transfer at 1:50 and continue to culture to OD 600 1.2-1.8; then centrifuge at 5000rpm for 5min, discard the supernatant, resuspend with the transformation medium, and adjust to the OD of the ba...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com