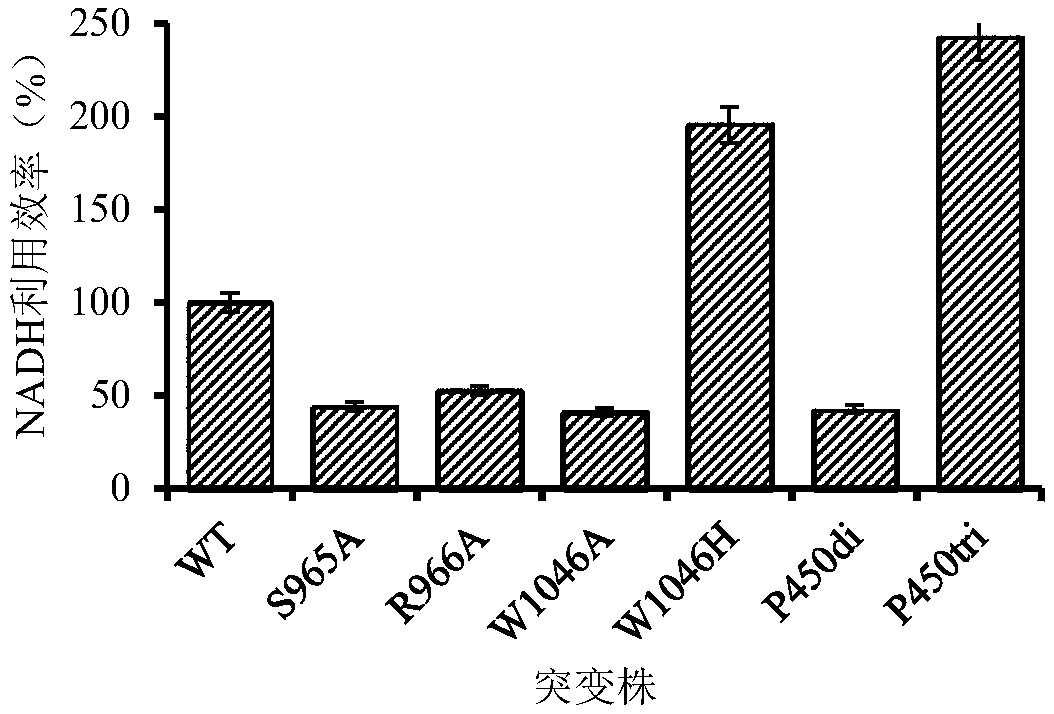Mutation-modified bacillus megaterium ALA2 cytochrome P450 enzyme, and preparation method and application thereof
A technology of Bacillus megaterium and cytochrome, applied in the field of molecular biology, can solve problems such as unfavorable flow to FAD, affect enzyme catalytic efficiency, reduce electron transfer efficiency, etc., and achieve the effect of increasing utilization efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Embodiment 1: Analysis and directional transformation of Bacillus megaterium P450 gene
[0021] 1.1 Determination of mutation sites
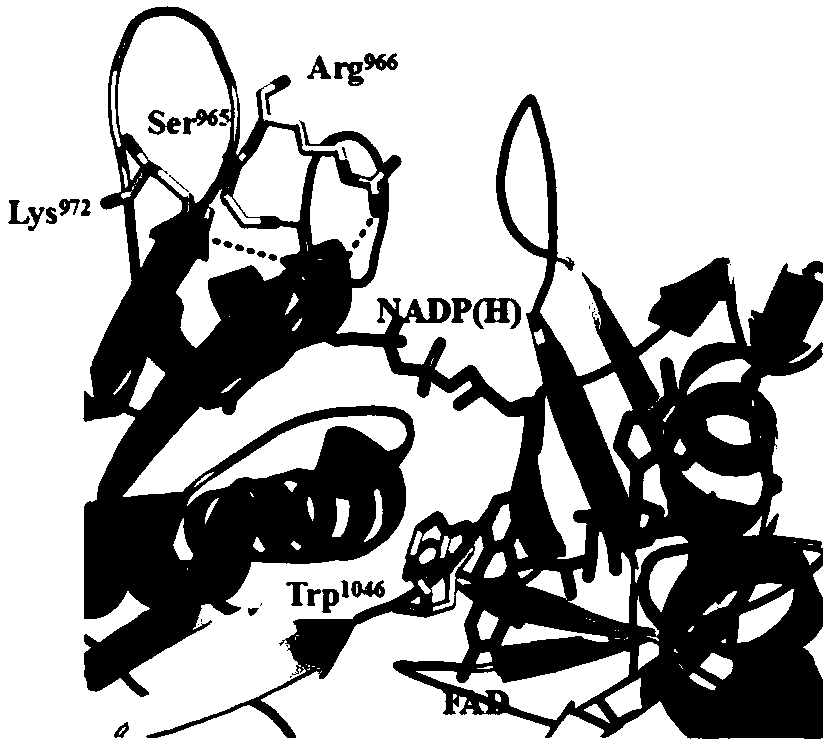
[0022] Directed Modification of Cytochrome P450 Enzyme Gene of Bacillus megaterium
[0023] Submit cytochrome P450 via SWISS-MODEL server (http: / / swissmodel.expasy.org / ) BM-3 Amino acid sequence information for the enzyme monooxygenase, resulting in cytochrome P450 BM-3 Among the locally resolved structures of the enzyme monooxygenase, its NADPH-P450 reductase flavin-binding domain and NADPH complex crystals have good resolution ( PDB ID: 4DQL), can be used for the analysis of coenzyme specificity. With the help of Swiss-pdbViewer4.03 software (Arnold K, Bordoli L, Kopp J T. The SWISS-MODEL workspace: a web-basedenvironment for protein structure homology modeling [J]. Bioinformatics, 2006, 22 (2): 195 -201.; Schwede T, Kopp J, Guex N, et al. SWISS-MODEL: An automated proteinhomology-modeling server. [J]. Nucleic Acids Research, 2003,...
Embodiment 2
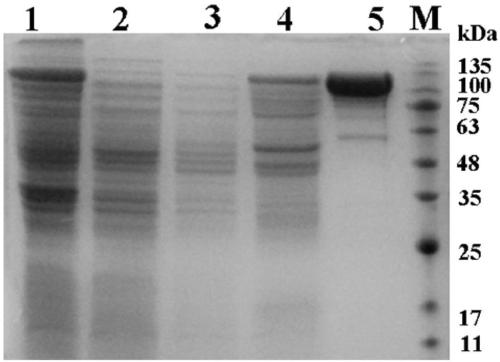
[0044] Embodiment 2: the preparation of recombinant enzyme and the mensuration of enzymatic activity
[0045] 2.1 Preparation of recombinant enzyme
[0046] Since the recombinant plasmid pTrc99A-cypt contains a His-tag tag, it was purified by His·Bind Purification Kit (Novagen) to obtain a purified recombinant enzyme. Specific operation process:
[0047] A. Processing of samples
[0048] (1) Resuspend the washed bacteria with 8 mL of 1×Binding Buffer, and break the wall by ultrasonic.
[0049] (2) After breaking the wall, centrifuge at 13,000 g for 30 min, and take the supernatant as the sample.
[0050] B. Handling Columns
[0051] (1) Take 1mL packing material and pack it into the column.
[0052] (2) Wash the column with 3 mL of sterile water.
[0053] (3) Wash the column with 5mL of 1×Charge Buffer.
[0054] (4) Wash the column with 3mL of 1×Binding Buffer.
[0055] C. Loading
[0056] (1) Add the sample to the column and control the flow rate to about 6 drops per...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com