Nucleic acid aptamer for detecting bladder cancer and its application
A nucleic acid aptamer and bladder cancer technology, applied in the field of tumor molecular biology, can solve the problems of high antibody immunogenicity, damage, systemic infection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1 The nucleic acid aptamer was screened by cell-SELEX technology.
[0055] (1) Design of the nucleic acid library and primers used:
[0056] Random ssDNA library:
[0057] 5'-CGTACGGTCGACGCTAGCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCACGTGGAGCTCGGATCC-3' (N represents four arbitrary bases of A, T, G and C) (SEQ NO.8)
[0058] Upstream primer: 5'-fluorescein isothiocyanate-CGTACGGTCGACGCTAGC-3'(SEQ NO.9)
[0059] Downstream primer: 5'-biotin-GGATCCGAGCTCCACGTG-3' (SEQ NO.10)
[0060] (2) Screening process:
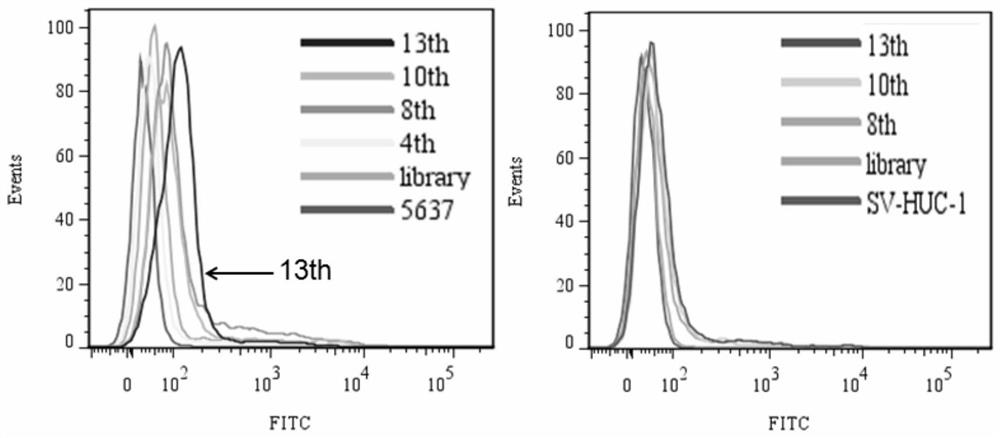
[0061] In the present invention, human bladder transitional cell carcinoma cell 5637 is used as a positive screening target, and human bladder urothelial cell SV-HUC-1 is used as a reverse screening target.
[0062] 2.1 Positive screening:
[0063] a. Incubation: Dissolve the above random DNA library with binding buffer, denature at 95°C for 5 minutes, refold on ice for 10 minutes, and then combine with human bladder transitional cell carcinoma cell 5637, whic...
Embodiment 2
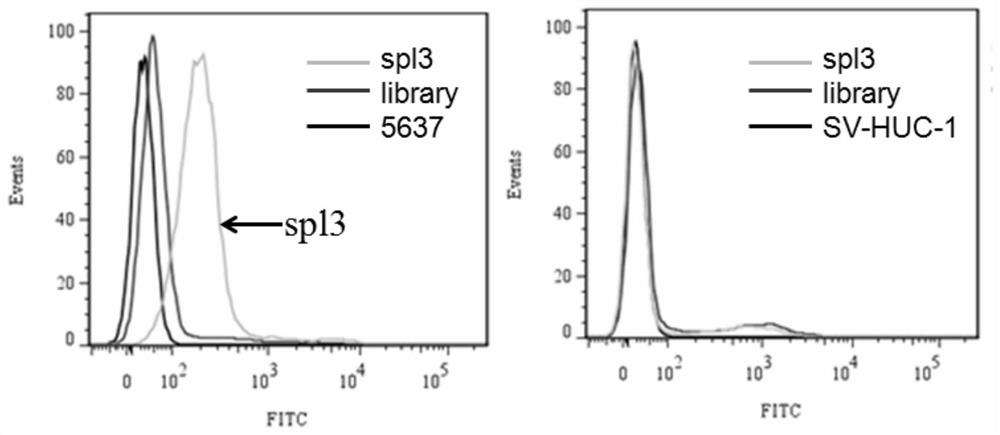
[0070] Example 2 Flow cytometric detection of the binding of the nucleic acid aptamer spl3 to human transitional bladder cancer cell 5637 and normal bladder urothelial cells SV-HUC-1
[0071] First, 5637 cells and SV-HUC-1 cells were cultured for 24 hours to make the cell density reach 90%, and then the adherent cells were digested from the culture dish with 0.2% EDTA respectively. Prepare 250nM synthetic FAM-labeled spl3 with 200μl of binding buffer, denature at 95°C for 5min, and refold on ice for 10min. Incubate with 300,000 5637 cells or SV-HUC-1 cells at 4°C for 1 hour. Wash the incubated cells 2-3 times with wash buffer, then resuspend the cells in 300 μl wash buffer. Fluorescent detection was performed by flow cytometry, and an initial random library of DNA was used as a control. The nucleic acid aptamer only binds to the target cell 5637, but not to the normal bladder urothelial cell SV-HUC-1. The results are as follows figure 2 shown.
Embodiment 3
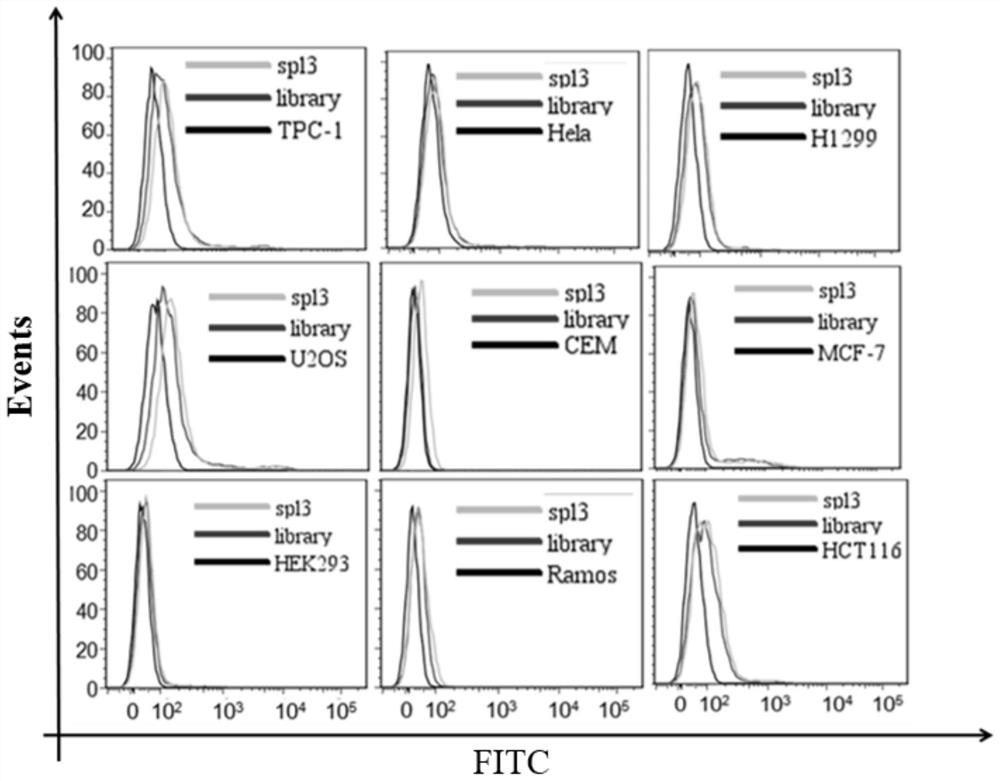
[0072] Example 3 The binding of the obtained nucleic acid aptamer spl3 to other solid tumor cancer cells was detected by flow cytometry.
[0073] Firstly, different types of cells were cultured for 24 hours to make the cell density reach 90%, and then the adherent cells were digested from the culture dish with 0.2% EDTA respectively. Prepare 250nM synthetic FAM-labeled spl3 with 200μl of binding buffer, denature at 95°C for 5min, and refold on ice for 10min. Incubate with 300,000 cells for 1 h at 4°C. Wash the incubated cells 2-3 times with wash buffer, then resuspend the cells in 300 μl wash buffer. Fluorescent detection was performed by flow cytometry, and an initial random library of DNA was used as a control. Nucleic acid aptamer spl3 does not bind to other solid tumor cells, the results are as follows image 3 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com