Nucleic acid aptamers for detecting bladder cancer and application thereof
A nucleic acid aptamer, bladder cancer technology, applied in the field of tumor molecular biology, can solve the problems of damage, high antibody immunogenicity, systemic infection, etc., achieve no cytotoxicity, high binding affinity and specificity, and improve the effect of affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
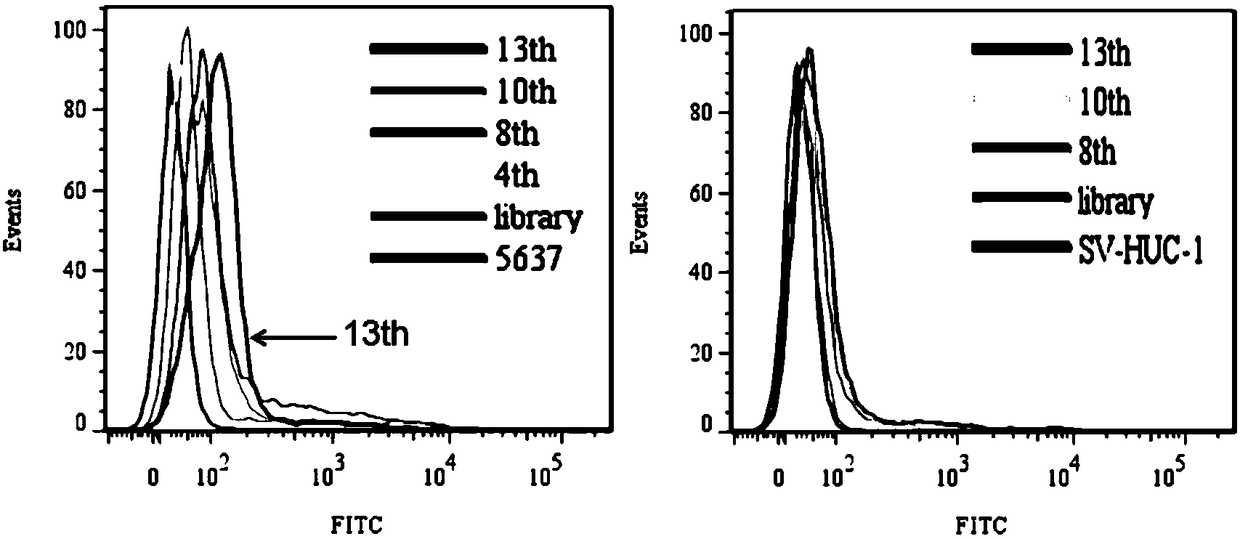
[0054] Example 1 The nucleic acid aptamer was screened by cell-SELEX technology.
[0055] (1) Design of the nucleic acid library and primers used:
[0056] Random ssDNA library:
[0057] 5'-CGTACGGTCGACGCTAGCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCACGTGGAGCTCGGATCC-3' (N represents four arbitrary bases of A, T, G and C) (SEQ NO.8)
[0058] Upstream primer: 5'-fluorescein isothiocyanate-CGTACGGTCGACGCTAGC-3'(SEQ NO.9)
[0059] Downstream primer: 5'-biotin-GGATCCGAGCTCCACGTG-3' (SEQ NO.10)
[0060] (2) Screening process:
[0061] In the present invention, human bladder transitional cell carcinoma cell 5637 is used as a positive screening target, and human bladder urothelial cell SV-HUC-1 is used as a reverse screening target.
[0062] 2.1 Positive screening:
[0063] a. Incubation: Dissolve the above random DNA library with binding buffer, denature at 95°C for 5 minutes, refold on ice for 10 minutes, and then combine with human bladder transitional cell carcinoma cell 5637, whic...
Embodiment 2
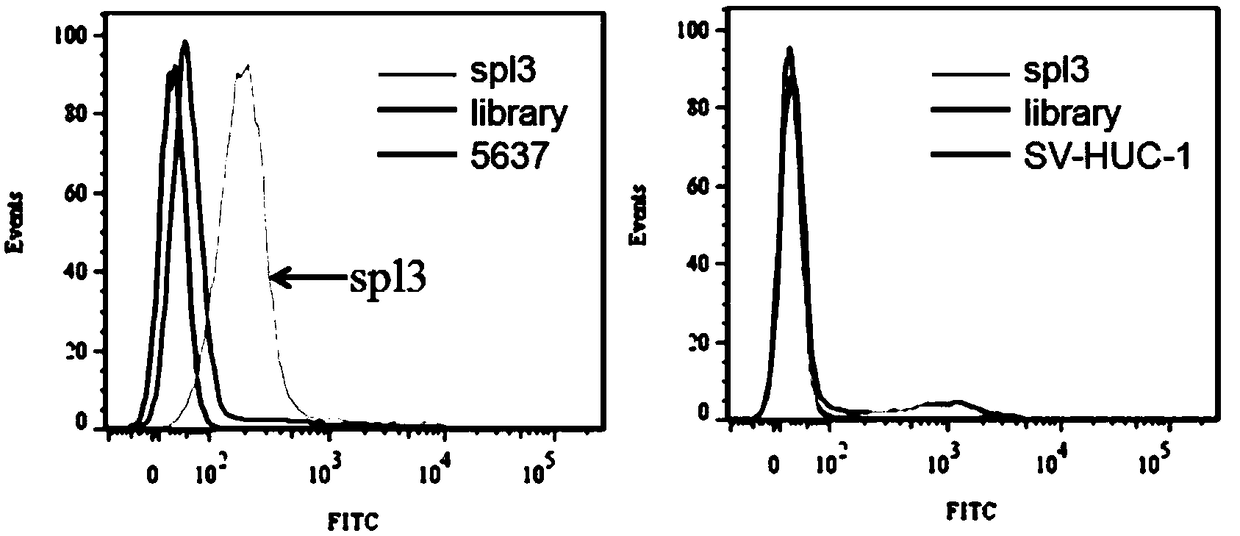
[0070] Example 2 Flow cytometric detection of the binding of the nucleic acid aptamer spl3 to human transitional bladder cancer cell 5637 and normal bladder urothelial cells SV-HUC-1
[0071] First, 5637 cells and SV-HUC-1 cells were cultured for 24 hours to make the cell density reach 90%, and then the adherent cells were digested from the culture dish with 0.2% EDTA respectively. Prepare 250nM synthetic FAM-labeled spl3 with 200μl of binding buffer, denature at 95°C for 5min, and refold on ice for 10min. Incubate with 300,000 5637 cells or SV-HUC-1 cells at 4°C for 1 hour. Wash the incubated cells 2-3 times with wash buffer, then resuspend the cells in 300 μl wash buffer. Fluorescent detection was performed by flow cytometry, and an initial random library of DNA was used as a control. The nucleic acid aptamer only binds to the target cell 5637, but not to the normal bladder urothelial cell SV-HUC-1. The results are as follows figure 2 shown.
Embodiment 3
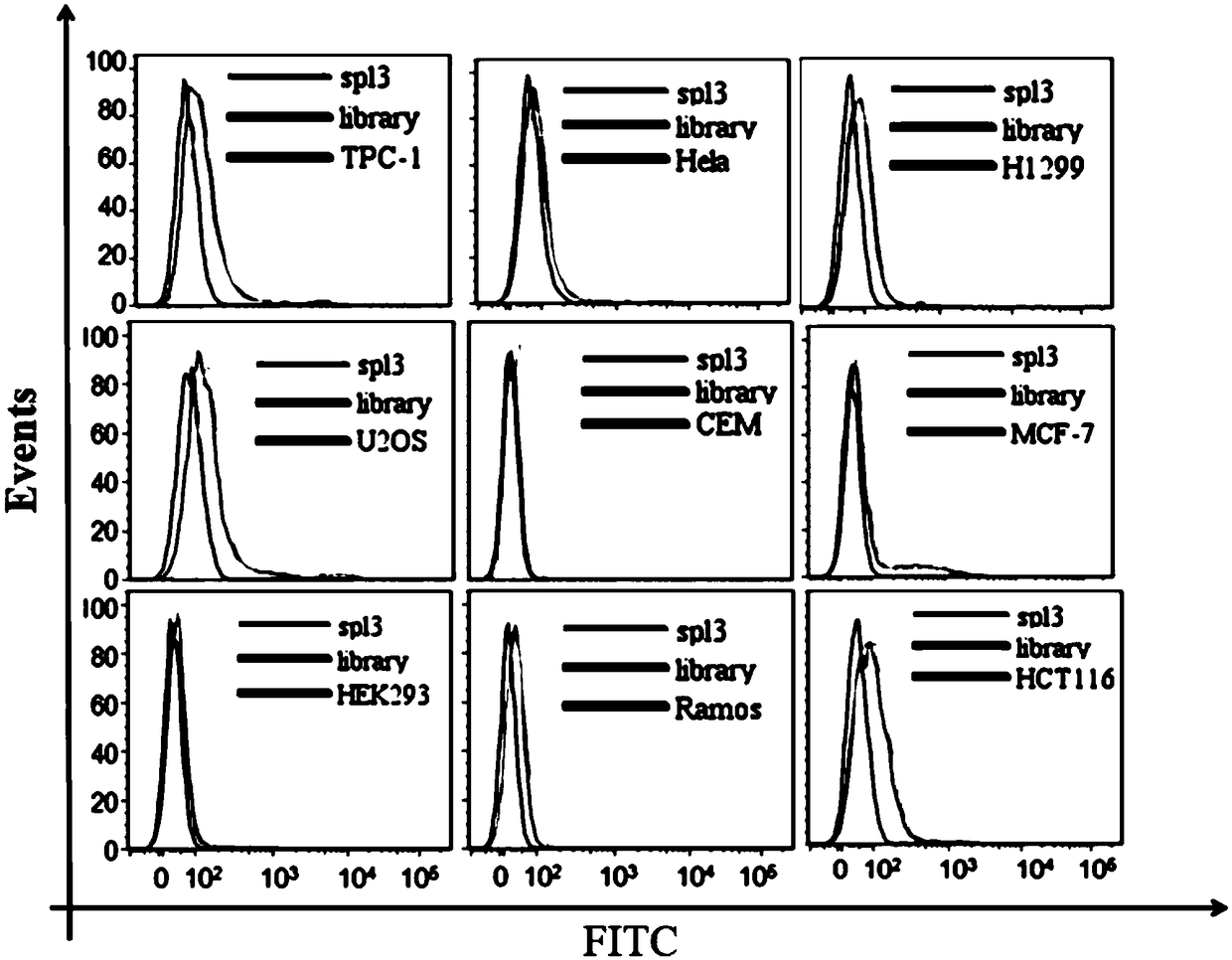
[0072] Example 3 The binding of the obtained nucleic acid aptamer spl3 to other solid tumor cancer cells was detected by flow cytometry.
[0073] Firstly, different types of cells were cultured for 24 hours to make the cell density reach 90%, and then the adherent cells were digested from the culture dish with 0.2% EDTA respectively. Prepare 250nM synthetic FAM-labeled spl3 with 200μl of binding buffer, denature at 95°C for 5min, and refold on ice for 10min. Incubate with 300,000 cells for 1 h at 4°C. Wash the incubated cells 2-3 times with wash buffer, then resuspend the cells in 300 μl wash buffer. Fluorescent detection was performed by flow cytometry, and an initial random library of DNA was used as a control. Nucleic acid aptamer spl3 does not bind to other solid tumor cells, the results are as follows image 3 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com