A molecular marker and its application for rapid detection of high-yielding genes in Thiopyrum elongatum
A technology of Thinpyrum and gene, which is applied in the rapid detection of molecular markers and application fields of high-yield genes of Thinopyrum elongatum, can solve the problems of lack of molecular markers of high-yield genes, difficulty in phenotypic identification, and low breeding efficiency, so as to improve breeding efficiency, Reduce the cost of breeding and reduce the effect of the operation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 Creation of Near-Isogenic Lines of High Yield Genes of Wheat-Thinopyrum elongatum
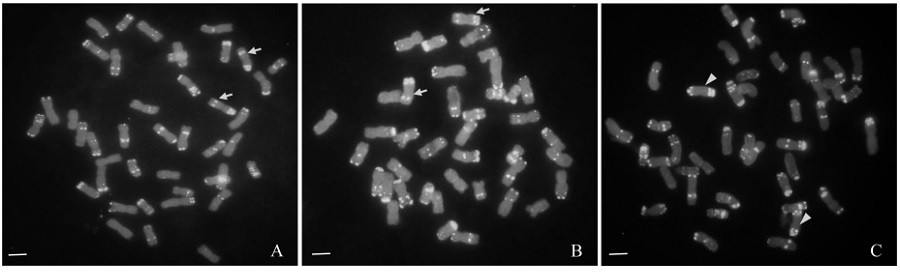
[0060] In 2015, the hybrid combination (Liangxing 66×SNK064) was configured in the greenhouse of the Crop Research Institute of Shandong Academy of Agricultural Sciences, and obtained F 1 ; Spring 2016, will receive F 1 Backcross with Liangxing 66 and get BC 1 f 1 , and carry out molecular marker identification on the obtained plants, on the basis of molecular marker identification, the BC carrying target gene and exogenous fragment 1 f 1 Continue to backcross with Liangxing 66 and get BC 2 f 1 . In the summer and autumn of 2016, the artificial climate chamber continued to add generations, self-crossed twice, and obtained BC 2 f 3 , with the help of molecular markers and fluorescence in situ hybridization, a wheat-Thinopyrum elongatum 7el2L heterozygous line was obtained; in the winter of 2016 and the spring of 2017, the above heterozygous line continued to be selfed twic...
Embodiment 2
[0062] Example 2 Analysis of agronomic and yield-related traits of high-yielding near-isogenic lines of Echinopsis elongatum
[0063] Refer to the method of Tang et al. After the in situ hybridization chromosome preparation was dried naturally, 0.25 μg / mL propidium iodide was added dropwise and covered with a cover glass, and photographed under an Olympus BX51 fluorescence microscope.
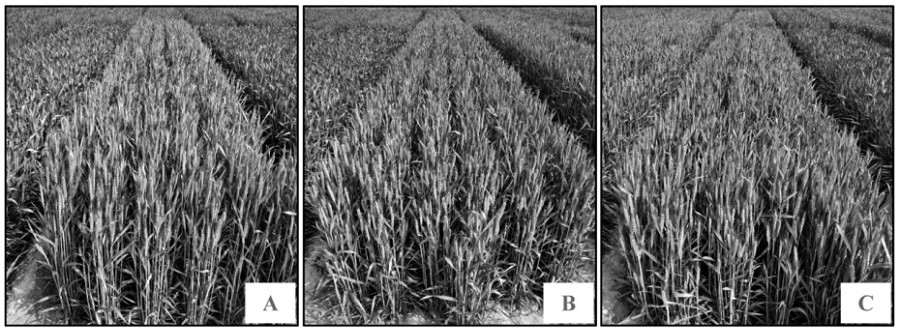
[0064] The above-mentioned near-isogenic line materials (SAAS1001, SAAS1002 and SAAS1003) were planted in the Lingcheng Experimental Base in the winter of 2017, with an area of 1.5m×6.0m, and repeated 3 times. For the flowering stage and grain filling stage of wheat, the length and width of the flag leaf, the length of the ear, the number of grains per ear, the number of fertile spikelets, the number of sterile spikelets, etc. 6 traits were investigated and analyzed, and 30 panicles were investigated in each plot. After harvesting, use the Wanshen SC-G automatic seed testing and thousand-gr...
Embodiment 3
[0066] Example 3 Development of Tightly Linked Molecular Markers for High Yield Genes of Echinopsis elongatum
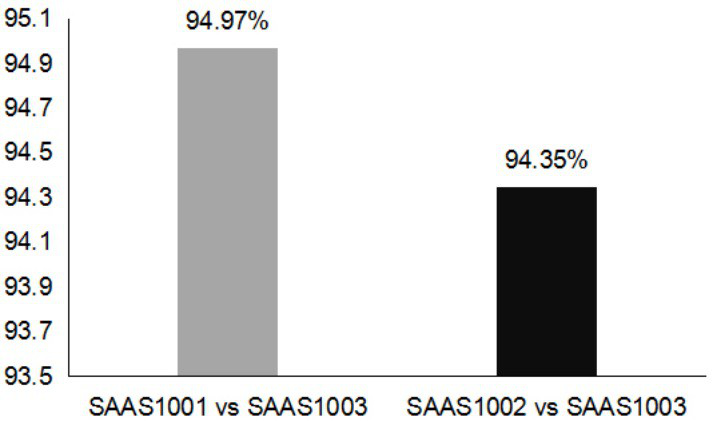
[0067] According to the sequence information of the published wheat genome (7DL end), SSR primers were designed using Primer 3.0, and wheat varieties Jimai 22, Liangxing 66, SAAS1001 and Jimai 22 mixed DNA, SAAS1002 and Liangxing 66 mixed DN, SAAS1003 and SAAS1001 Mixed DNA and wheat-Thinopyrum elongated near-isogenic lines SAAS1001, SAAS1002 and SAAS1003 were used as materials for PCR amplification to search for molecular markers closely linked to high-yielding genes.
[0068] In the PCR amplification reaction system, the volume of the PCR reaction system is 10.0 μL, including 10×Buffer buffer: 1.0 μL, 2.5 mM dNTP solution: 0.8 μL, Taq enzyme 0.1 μL, sterilized double distilled water: 4.1 μL, 50ng DNA Template 2.0 μL (Jimai 22, Liangxing 66, SAAS1001 and Jimai 22 mixed DNA, SAAS1002 and Liangxing 66 mixed DNA, SAAS1003 and SAAS1001 mixed DNA, and wheat-Ethiopia elon...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com