SNP molecular marker for biomphalaria and application of SNP molecular marker in traceability
A molecular marker, double umbilical snail technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragment, etc. Effects with small distribution differences and rich polymorphisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Search of SNP molecular markers
[0031] (1) Construction of DNA pool (pool)
[0032] Two umbilical snails in Shenzhen, Dongguan and Hong Kong were randomly collected respectively, and after the genomic DNA was extracted, the DNA of 6 individuals was aspirated and put into the same centrifuge tube, and mixed well for use.
[0033] (2) Primer design
[0034] Using the DNA sequence of the LOC106079843 gene of the umbilical snail (Genbank: NW_013487738.1) as a template, primers were designed to isolate the DNA fragment of the LOC106079843 gene of the umbilical snail. The primers are as follows:
[0035] Upstream primer: 5´-AAGAGTTGTGAGAGTGGGCC-3´ (as shown in SEQ ID NO.2)
[0036] Downstream primer: 5´-TCGGCCGAAAGTCATTATATGG-3´ (as shown in SEQ ID NO.3)
[0037] The total volume of the PCR reaction was 50 μL, including about 100 ng of A. umbilicus genomic DNA, 45 μL of 1.1×PCR mix, and 2 μL of 10 μmol / L upstream and downstream primers. The PCR amplification p...
Embodiment 2
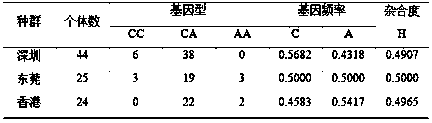
[0044] Example 2 Distribution of alleles
[0045] (1) Design of test groups
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com