Gene knockout method
A gene knockout and gene technology, applied in the field of gene editing, can solve problems such as low integration efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
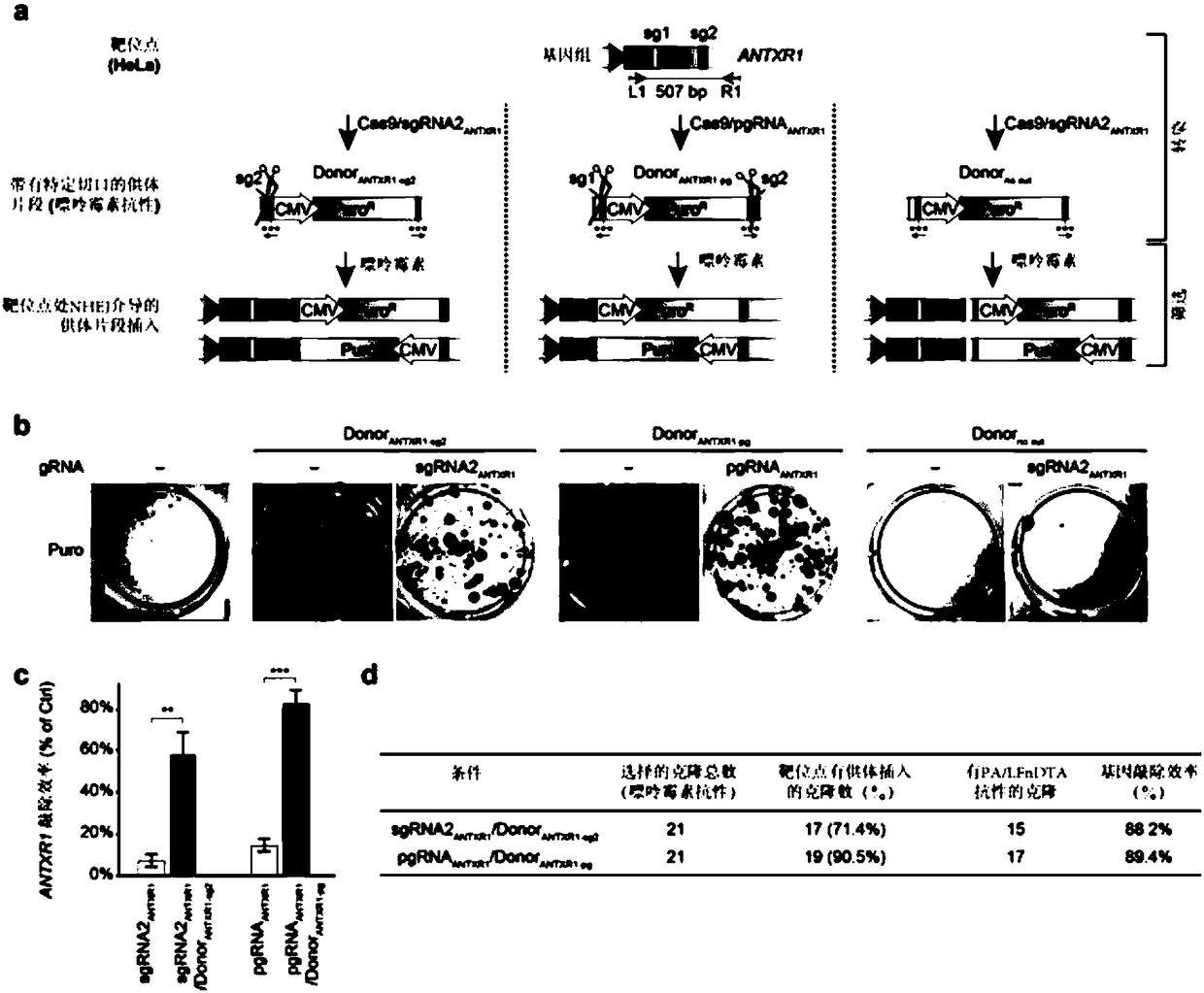
[0165] Example 1 Enrichment of Knockout Events on ANTXR1 Gene in HeLa Cells Using Linear Donor DNA
[0166] 1. Design of sgRNA
[0167] Two sgRNAs targeting the first exon of the ANTXR1 gene in HeLa cells were designed, and their efficiency in generating deletion or insertion mutations (Indels) at the target site was verified by T7E1 assay. The verification results are shown in Table 1. where sgRNA1 ANTXR1 The targeted sequence is referred to as sg1 in this example, sgRNA2 ANTXR1 The targeted sequence is referred to as sg2 in this example.
[0168] Table 1 sgRNA targeting the first exon of ANTXR1 gene in HeLa cells
[0169]
[0170] 2. Construction of Linear Donor DNA
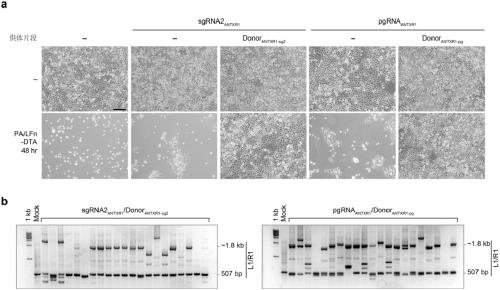
[0171] A total of two linear donor DNAs (Donor ANTXR1-sg2 and Donor ANTXR1-pg ), whose structure refers to figure 1 a.
[0172] Donor ANTXR1-sg2 From 5' to 3' ends include: 20bp protection sequence, sg2, reverse stop codon, CMV promoter-driven puromycin resistance gene, forward stop codon, 20bp pr...
Embodiment 2
[0182] Example 2 Enrichment of Knockout Events on the HBEGF Gene in HeLa Cells Using Linear Donor DNA
[0183] Since donors with either single- or double-cleavage sites can greatly improve the selection of cells with modifications at the target site, for convenience only single-cleavage donors were used in this example.
[0184] 1. Design of sgRNA
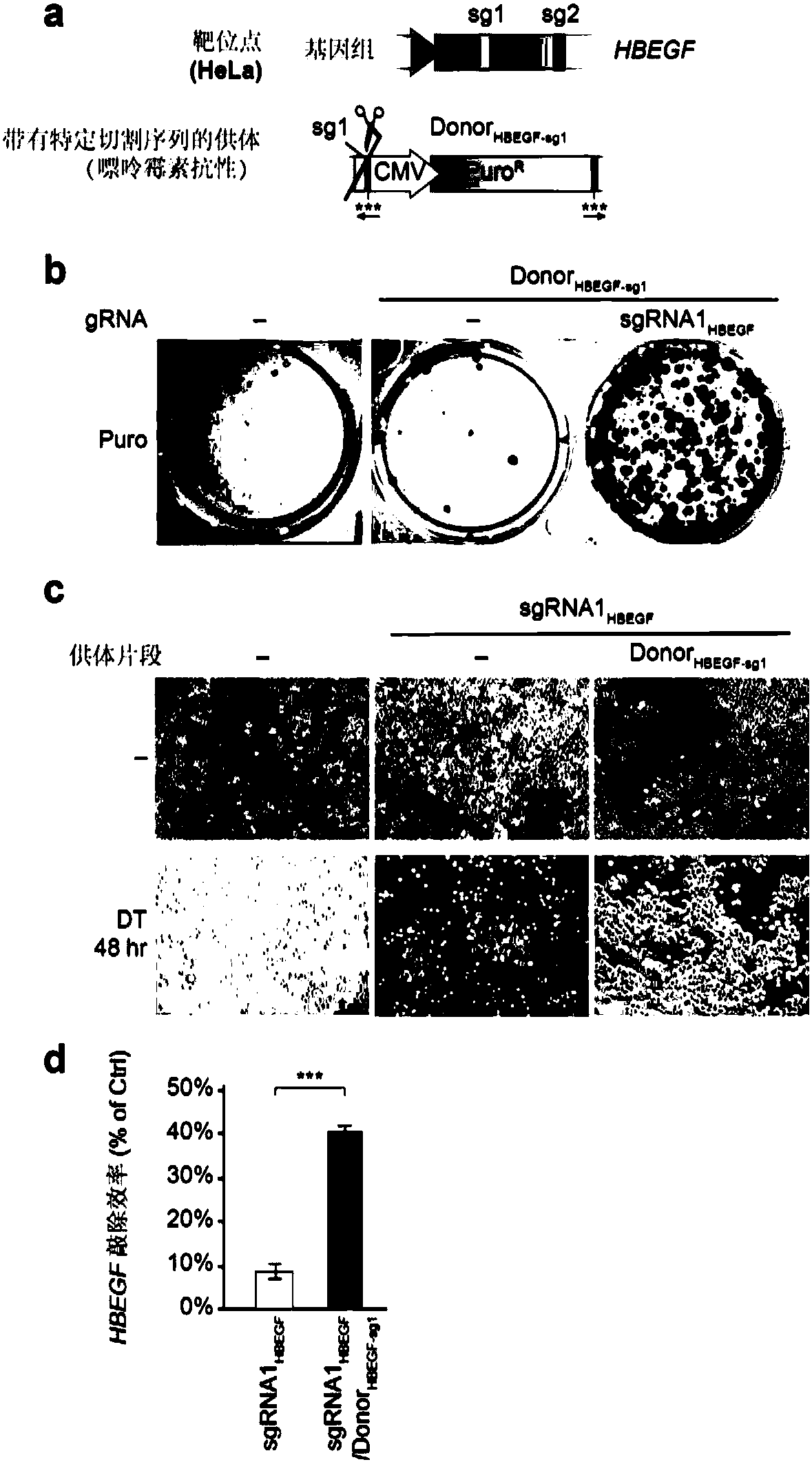
[0185] Two sgRNAs targeting the HBEGF gene in HeLa cells were designed, and their efficiency of generating Indels at the target site was verified by T7E1 assay. The verification results are shown in Table 3. where sgRNA1 HBEGF The target sequence against is called sg1 in this example, sgRNA2 HBEGF The targeted sequence is referred to as sg2 in this example.
[0186] Table 3 sgRNAs targeting the HBEGF gene in HeLa cells
[0187]
[0188]
[0189] 2. Construction of Linear Donor DNA
[0190] Construct a linear donor DNA (Donor HBEGF-sg1 ), whose structure refers to image 3 a.
[0191]Donor HBEGF-sg1 From 5' to 3' en...
Embodiment 3
[0197] Example 3 Enrichment of Knockout Events on HBEGF Gene in HEK293T Cells Using Linear Donor DNA
[0198] 1. Design of sgRNA and construction of linear donor DNA
[0199] Design of sgRNA2 targeting HBEGF gene in HEK293T cells HBEGF , and construct a linear donor DNA (Donor HBEGF-sg2 ), the donor includes: 20bp protection sequence from 5' to 3' end, sg2, reverse stop codon, EGFP gene driven by CMV promoter, forward stop codon, 20bp protection sequence, see Figure 4 a.
[0200] 2. Verification of gene knockout efficiency
[0201] Using a plasmid expressing Cas9 and sgRNA2 HBEGF , with or without its corresponding donor Donor HBEGF-sg2 , co-transfected HEK293T cells, cells were screened by FACS, EGFP-positive cells were screened by FACS in the group with donor, and mCherry-positive cells were screened by FACS in the group without donor. FACS-selected cells were treated with DT (40 ng / ml) to compare the effect of linear donor DNA on HBEGF knockdown efficiency. The im...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com