A dna methylation quantitative system
A methylation and non-methylation technology, applied in the field of analysis, can solve the problems of inability to accurately detect genomic heterozygous methylation, inability to detect methylation levels in regions other than CpG islands, and inability to successfully design primers and probes. , to achieve the effect of increasing the annealing temperature
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] Embodiment 1 A kind of DNA methylation quantitative system
[0103] A DNA methylation quantitative system comprising:
[0104] a) Collection component: used to collect the amount of methylation and the amount of non-methylation of the DNA sample to be tested;
[0105] Specifically, the collection component also includes:
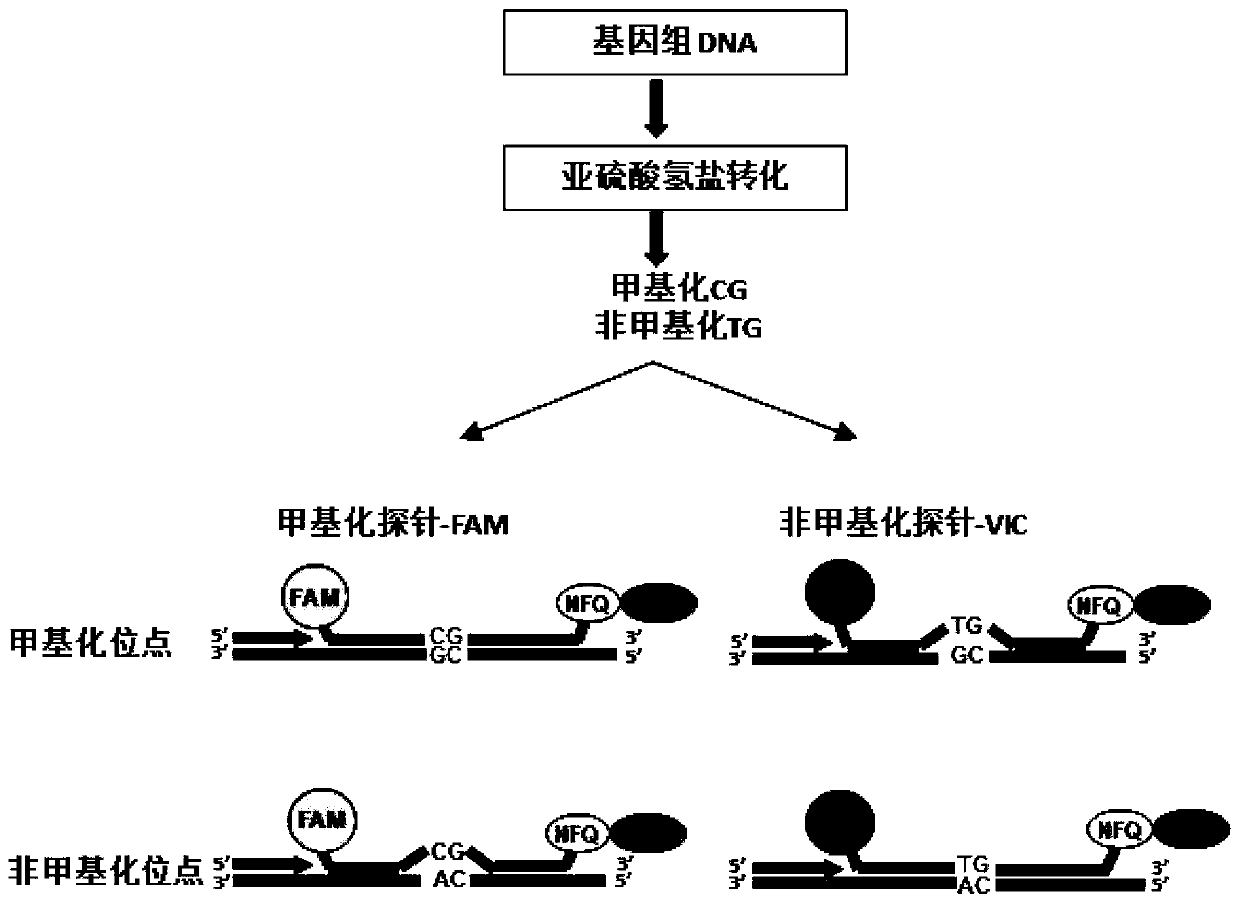
[0106] 1) conversion module: the conversion module is used to convert the unmethylated cytosine base of the DNA sample into uracil, while the methylated cytosine base remains unchanged;
[0107] 2) Amplification module: the amplification module is used to amplify the transformed DNA sample.
[0108] b) Processing components: methylation / (methylation+non-methylation)×100 to calculate the percentage of methylated referenc, PMR.
[0109] As an option, it also contains
[0110] c) Output component: the output component is used to output the processing component to obtain the methylation percentage parameter percentof methylated referenc, PMR.
Embodiment 2
[0111] Example 2 DNA methylation quantification system workflow
[0112] 1. The amount of methylation and non-methylation of DNA samples is collected
[0113] Material
[0114] Fresh-frozen tumor tissue samples were obtained from 45 patients with primary colorectal adenocarcinoma. Patients included 32 men and 13 women. Among these patients, 17 had stage I tumors and 28 had stage II tumors; 18 had relapsed and 27 had no recurrence. See Table 1 for details.
[0115] Table 1 Tumor samples
[0116]
[0117]
[0118] 2. The conversion module converts the unmethylated cytosine bases of the DNA sample to be tested
[0119] Using the QIAamp DNA Mini Kit (Qiagen, 51306) and the EZ DNA Methylation Kit (ZymoResearch, D5002), the genomic DNA in the above samples was extracted and modified with bisulfite according to the instructions [7,14].
[0120] 3. The amplification module amplifies the transformed DNA sample
[0121] After bisulfite conversion, real-time fluorescent quan...
Embodiment 3
[0147] Example 3 Bisulfite Pyrosequencing Comparative Evaluation of Quantitative Accuracy
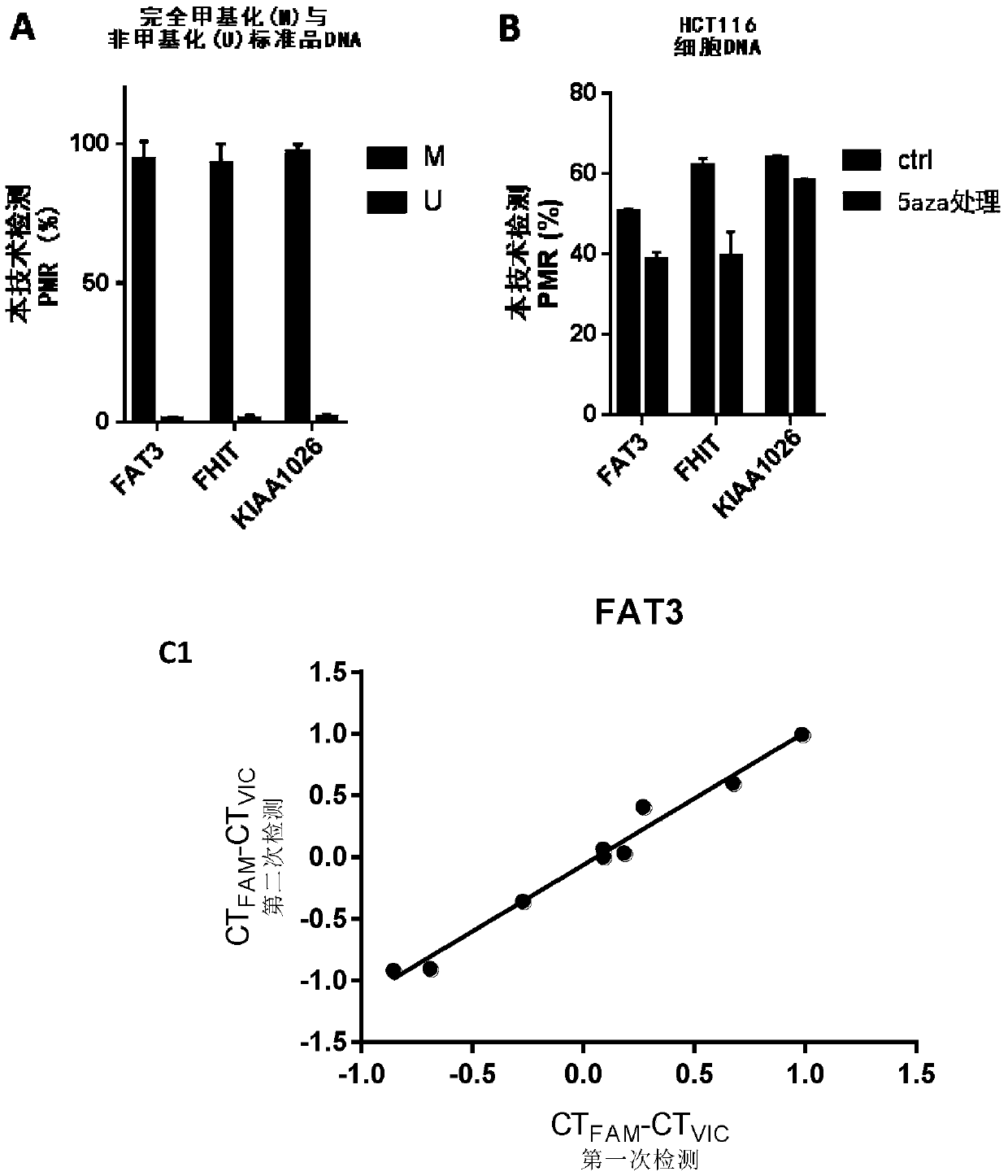
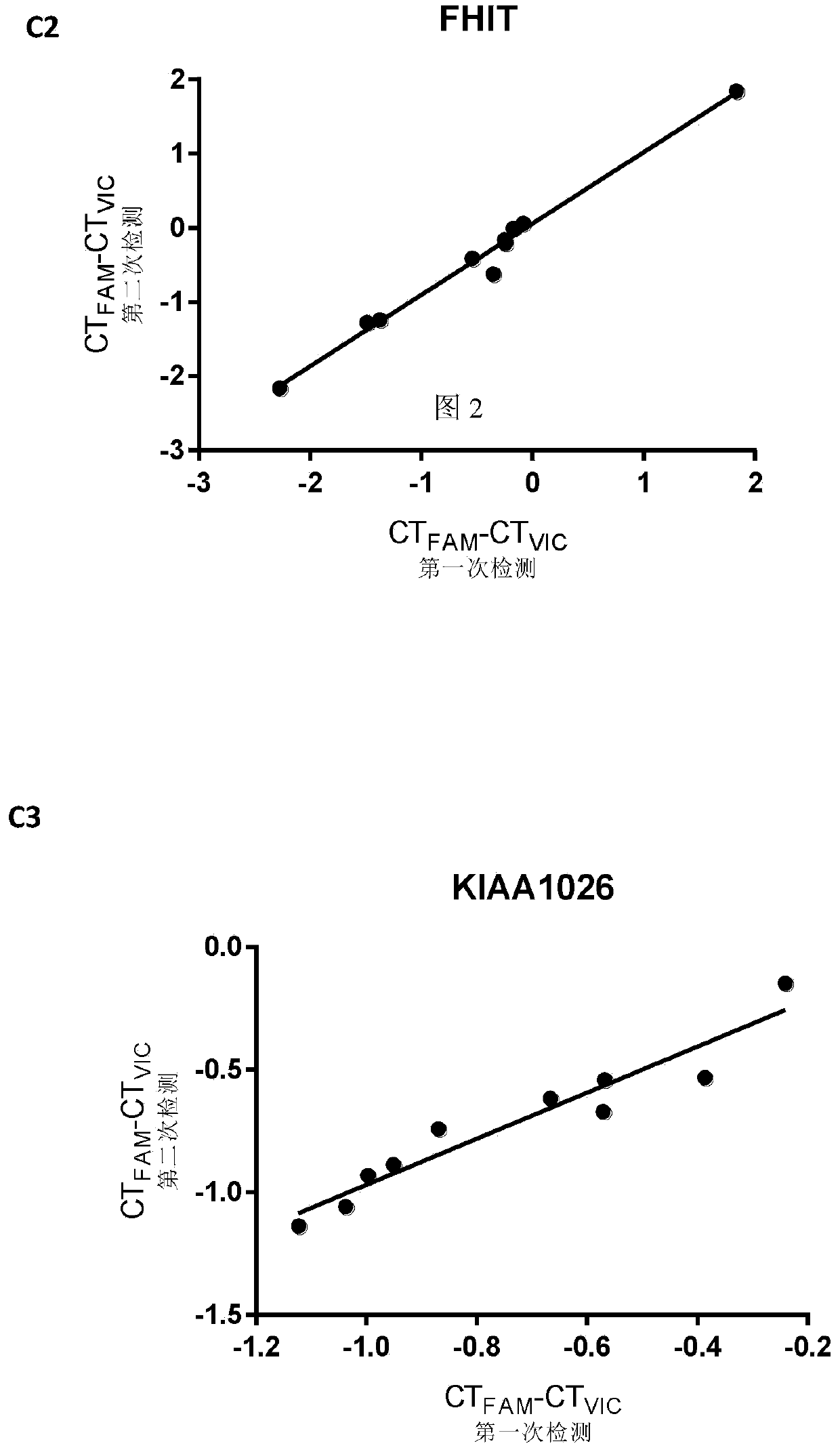
[0148] Using pyrosequencing as a reference, the accuracy of the methylation quantification system of the present invention was further tested. We used pyrosequencing and this technology to detect the methylation levels of the above three CpG sites in 10 colorectal cancer tissues (Table 1). The primers for pyrosequencing are listed in Table 2. The PMR measured by the present invention is linearly correlated with the methylation percentage obtained by pyrosequencing ( image 3 ; FAT3 was 0.9690, FHIT was 0.9954, KIAA1026 was 0.8755, all P<0.001). Not only that, there is a striking agreement between PMR and Pyrosequencing in terms of percent methylation. This is more precise than traditional MethyLight reported previously [11,12]. Therefore, this system has the same quantitative accuracy as bisulfite pyrosequencing, but is easier to operate, more accessible, and less expensive.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com