Quantitative fluorescent PCR inspection for hepatitis-B virus drug-tolerant gene mutation
A hepatitis B virus and detection method technology, applied in the field of fluorescent quantitative landing PCR detection, can solve the problems of poor accuracy, low detection sensitivity, and bottlenecks in the detection of mixed infected strains, achieve accurate and rapid detection, and reduce non-specific amplification Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
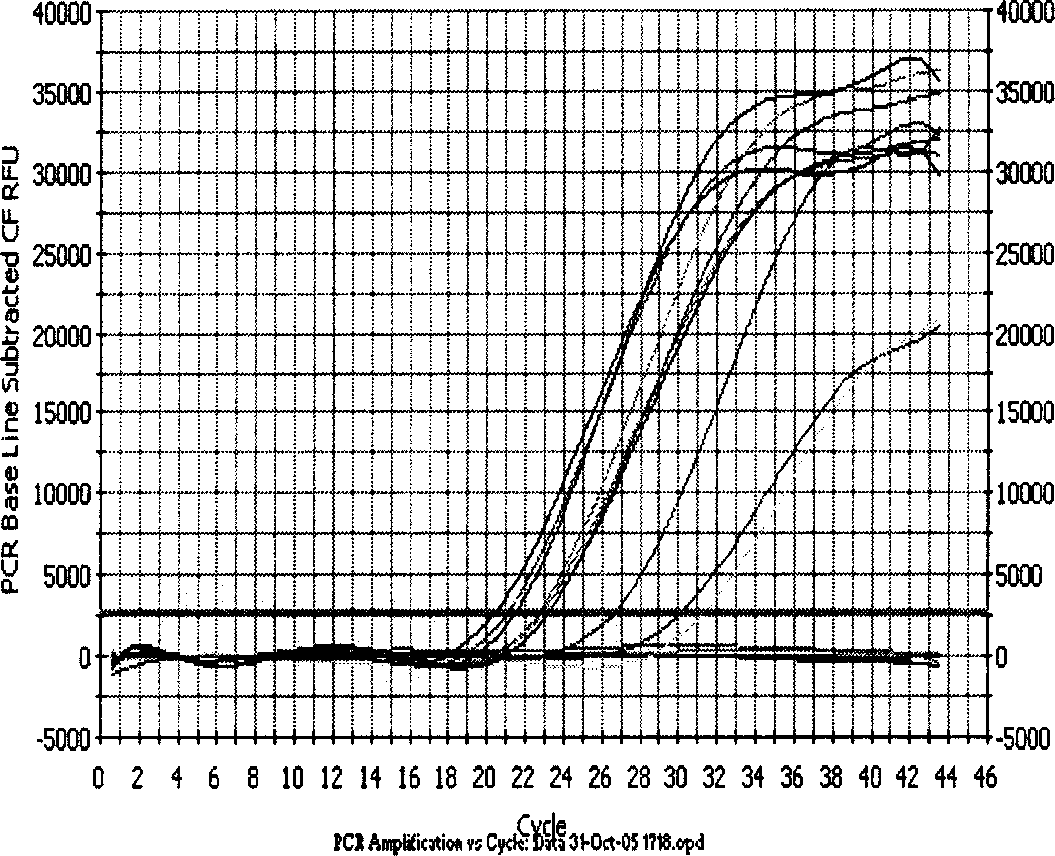
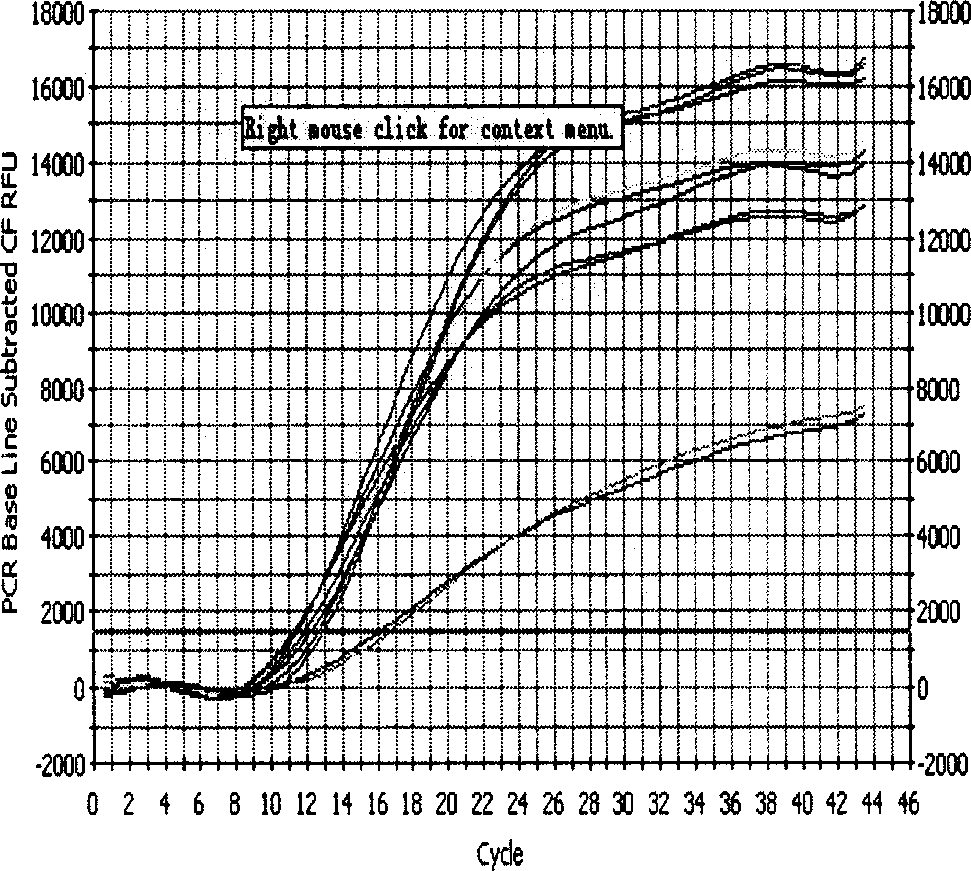
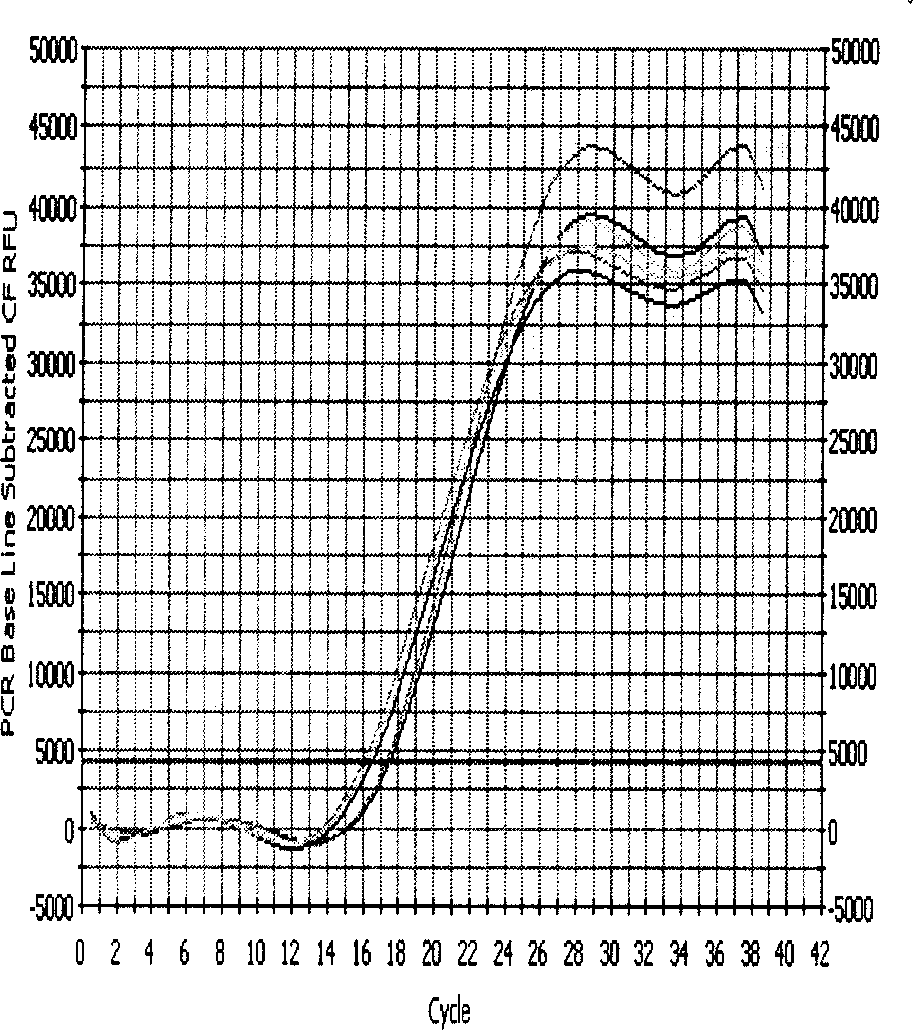
[0020] 1. Design and synthesis of primers for lamivudine-resistant gene mutation of hepatitis B virus fluorescent quantitative PCR
[0021]Using Beacon Designer 2.1 (molecular beacon design software 2.1) to design fluorescence quantitative PCR lamivudine resistance gene mutation primers. Among them, the mutant and wild sites are located at the 3' end of the downstream primer, and the downstream primers corresponding to the M204 wild-type primer in the polymerase C functional region and the mutant M204V and M204I are YMDD-L, YVDD-L, and YIDD-L, respectively; The downstream primers of L180 in the polymerase B functional region and the mutated L180M are 180W-L and 180M-L, respectively. The upstream primer PU is located in the conserved region of the polymerase and is used to pair with the 5 downstream primers. Among them, the size of the PCR product obtained by pairing PU with 180W-L and 180M-L is 360BP (Base Pair, base pair), and the size of the PCR product obtained by pairing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com