Molecular subtyping method for salmonella typhimuria and specific SNP locus combination
A Salmonella and molecular typing technology, applied in the fields of biochemical equipment and methods, recombinant DNA technology, and microbial determination/inspection, etc., which can solve the problems of incomplete data sources, small amount of data, and lack of strain data.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
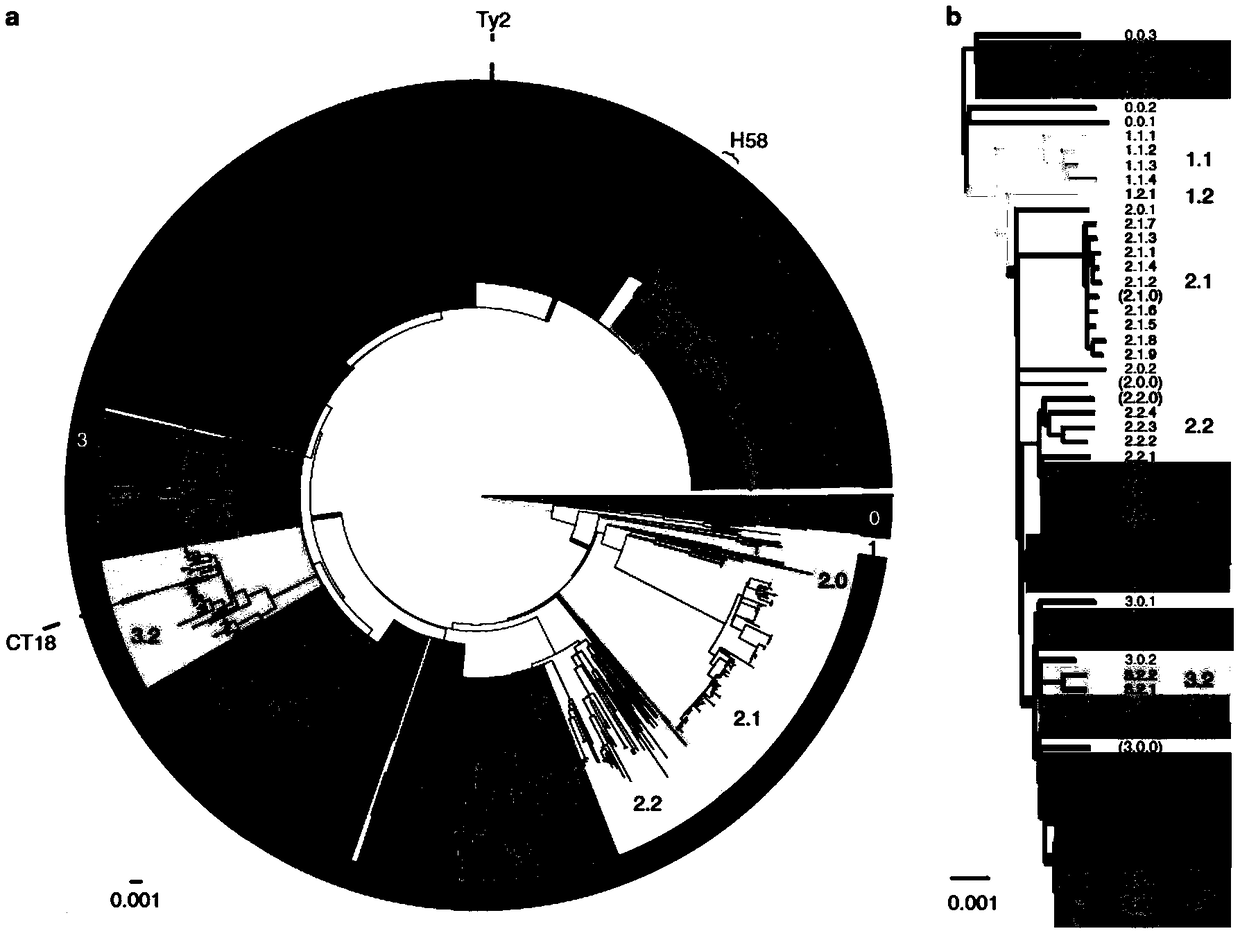
[0102] Example 1. Molecular typing of Salmonella typhimurium worldwide and screening of specific SNP site combinations
[0103] Flow chart of molecular typing of Salmonella typhimurium figure 2 As shown, the specific process is as follows:
[0104] 1. Get data
[0105] The whole genome sequencing data of 6637 strains of Salmonella typhimurium worldwide were obtained from http: / / ncbi.nlm.nih.gov / , and the data format was processed into fastq or fasta format; Salmonella typhi DNA samples were sequenced by BGI to obtain data in fastq format.
[0106] 2. Verify the serotype
[0107] The whole genome sequencing data of Salmonella typhimurium strains obtained in the above 1 were used to predict the serotype by SeqSero software, and the data with inconsistent predicted types and phenotypes were screened out, and the whole genome sequencing data of global Salmonella typhimurium strains with consistent predicted types and phenotypes were retained. The global strain whole genome se...
Embodiment 2、4151
[0193] Example 2, 4151 strains of Salmonella typhimurium from Asia, Africa and Europe public database sequencing data verification method
[0194] The method of Example 1 was verified by using 4151 strains of Salmonella typhimurium public database sequencing data from Asia, Africa and Europe, specifically as follows:
[0195] 1) Perform FASTQ format conversion on the whole genome sequencing data of 4151 strains of Salmonella typhimurium from Asia, Africa and Europe from SRA data;
[0196] 2) Predict the serotype of the sample data, and eliminate the data inconsistent with the phenotype;
[0197] 3) Using the typhoid murine standard strain (accession: NC_016810) as the reference sequence, perform a variation search to find out all the SNP sites that are different from the typhoid murine standard strain (accession: NC_016810);
[0198]4) Retrieve the polymorphic forms of all the above SNP sites and the 15 groups of specific SNP site combinations (column 3 in Table 1) shown in T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com